Significance
Previous studies on the interaction between nucleoporins and HIV-1 capsid focused on the classical FG-motif and CA hexamer. In this study, we found that besides the FG-motif engaging the CA hexamer, a triple-arginine (RRR) motif at the C terminus of NUP153 targeted HIV-1 capsid at the interface of three CA hexamers. Both FG- and RRR-motifs were important for the nuclear import of HIV-1 cores and the infectivity of the virus. Moreover, NUP153 stabilized tubular CA assemblies in vitro. These results provide insights into how NUP153 contributes to both facilitating and maintaining the assembled capsid’s passage through the nuclear pore complex.
Keywords: NUP153, RRR-motif, HIV-1 capsid, lattice stabilization, nuclear entry
Abstract
Increasing evidence has suggested that the HIV-1 capsid enters the nucleus in a largely assembled, intact form. However, not much is known about how the cone-shaped capsid interacts with the nucleoporins (NUPs) in the nuclear pore for crossing the nuclear pore complex. Here, we elucidate how NUP153 binds HIV-1 capsid by engaging the assembled capsid protein (CA) lattice. A bipartite motif containing both canonical and noncanonical interaction modules was identified at the C-terminal tail region of NUP153. The canonical cargo-targeting phenylalanine-glycine (FG) motif engaged the CA hexamer. By contrast, a previously unidentified triple-arginine (RRR) motif in NUP153 targeted HIV-1 capsid at the CA tri-hexamer interface in the capsid. HIV-1 infection studies indicated that both FG- and RRR-motifs were important for the nuclear import of HIV-1 cores. Moreover, the presence of NUP153 stabilized tubular CA assemblies in vitro. Our results provide molecular-level mechanistic evidence that NUP153 contributes to the entry of the intact capsid into the nucleus.
As a retrovirus, HIV-1 reverse transcribes its RNA genome for integration into host chromatin in the nucleus (1). The HIV-1 genome is housed inside the viral capsid, which provides an enclosed compartment necessary for reverse transcription and concomitantly protects the genome from destruction by host immune surveillance and cellular antiviral restriction factors (2, 3). The capsid is composed of capsomeres of approximately 250 capsid protein (CA) hexamers and 12 CA pentamers, which assemble into a fullerene cone (4–6). To understand the life cycle of HIV-1, it is important to know whether and how HIV-1 capsid crosses the nuclear envelope and gains access to the nuclear compartment for the delivery of the viral genome. Previously, it was commonly thought that an intact capsid (6) was larger than the diameter of the nuclear pore complex (NPC) central transport channel (7); therefore, the capsid would need to uncoat in the cytosol prior to nuclear entry. However, compelling recent studies suggest that HIV-1 capsid can cross the NPC largely intact and disassembles only once inside the nucleus (8–10). However, the molecular mechanisms of capsid passage through the NPC have not been elucidated.
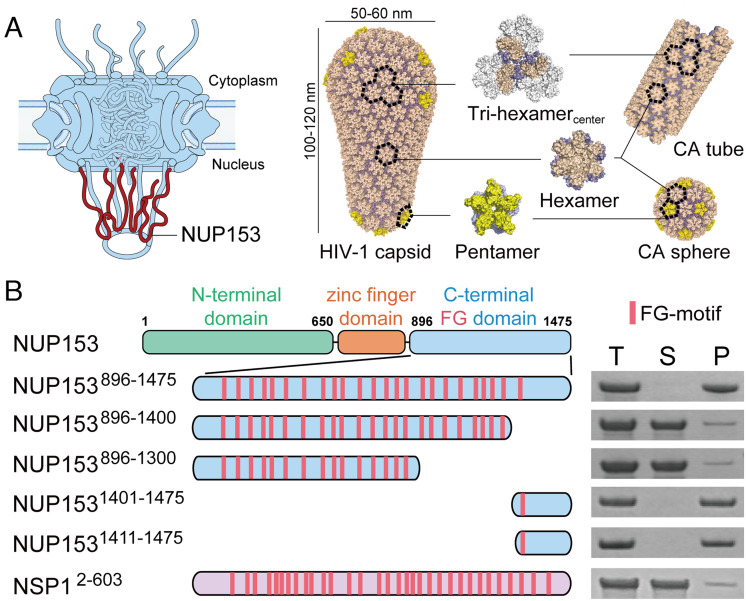
The NPC, which is among the largest macromolecular complexes in the human cell, is built from ~30 different nucleoporins (NUPs), each in 16 to 48 copies (11, 12). The NPC central channel is filled with intrinsically disordered NUPs rich in repeating phenylalanine-glycine (FG) motifs (11, 13). These FG-NUPs establish a selective barrier dictating the passage of macromolecules through the nuclear pore (14, 15). Many FG-NUPs have been reported to interact with the HIV-1 capsid, including NUP358, NUP214, NUP62, and NUP153 (16–21). Among these players, NUP153 is of particular interest. NUP153 is a component of the NPC’s nuclear basket (22, 23) and thus resides on the nuclear side of the NPC, consistent with its important role in the late stage of capsid nuclear transport (19, 24). Therefore, understanding its interaction with the capsid is key to unraveling the nuclear entry mechanisms of HIV-1, especially for the penetration of the capsid into, or its passage through, the NPC (Fig. 1A). NUP153 is a 1475 amino acid (aa)-long protein that contains 24 FG-repeats in its C-terminal domain (CTD) (Fig. 1B) (24, 25). Genetic analyses mapped a primary NUP153 interaction determinant to the most C-terminal FG-motif (24, 26), which through structural studies was shown to engage the interface between two adjacent CA subunits in a hexamer, albeit at a low binding affinity (Kd of ~50 to 500 μM) (21, 27). However, there is no clear indication whether this comparatively weak binding affinity is sufficient for the nuclear import of the capsid, and it remains unclear whether the virus–host interaction is enhanced by multivalent interactions, e.g., between additional NUP153 FG-motifs and higher-order capsid assemblies that extend beyond the hexamer building block.
Fig. 1.
The C-terminal 65 aa region of NUP153 mediates binding to CA assemblies. (A) Schematic of the NPC with relative NUP153 placement (Left) and various in vitro assembled CA structures used in this study (Right). (B) Copelleting assay of NUP153CTD variants and yeast NUP NSP1 control with CA nanotubes. Left: Schematics of the constructs with FG-motifs in each protein indicated as red bars. Right: The Total (T), Soluble (S), and Pellet (P) fractions of copelleting assays analyzed by SDS-PAGE. Independent experiments were repeated more than three times, with representative examples shown.
In this work, we used capsid engineering and mutagenesis techniques to map the interactions between various assembled CA interface modules and NUP153 (Fig. 1A). The results revealed a lattice pattern sensing ability of NUP153 that targets the assembled capsid over individual CA molecules or hexamers. A bipartite capsid-binding motif resides in the C-terminal 65 aa of NUP153, which recognizes the HIV-1 capsid via the canonical hexamer-binding FG-motif and a previously unidentified triple-arginine (RRR) motif engaging the threefold symmetry interface formed between three adjoining CA hexamers. HIV-1 infection studies revealed that both FG- and RRR-motifs were important for the nuclear import of HIV-1 cores. The presence of NUP153 can also stabilize assembled CA tubes in vitro, pointing to the possibility that NUP153 helps protect the capsid’s integrity as it facilitates the core’s passage through the central channel of the NPC.
Results
The C-terminal Tail of NUP153 Is Responsible for Interaction with CA Assemblies.
Using a genetic approach based on restriction of HIV-1 infection mediated via an artificial tripartite motif (TRIM)-NUP153CTD fusion protein, we previously implicated the most C-terminal FG-motif of NUP153 in mediating the interaction with capsid (24). However, as the assembled HIV-1 capsid contains numerous FG-binding pockets that interact with FG-motifs from NUP153 (21), cleavage and polyadenylation specificity factor 6 (CPSF6) (28), and SEC24C (29), we here used direct protein interaction assays to more comprehensively assess the roles of the different NUP153 FG-repeats in HIV-1 capsid binding. We created various C-terminal truncations of human NUP153CTD (residues 896 to 1475; NUP153896–1475), to isolate different FG-motifs into different NUP153CTD segments (Fig. 1B). We used a solubility-enhancing maltose-binding protein (MBP)-small ubiquitin-like modifier (SUMO) tag to produce well-behaved NUP153CTD constructs, which were shown by size exclusion chromatography (SEC) as mainly monodispersed monomers in solution (SI Appendix, Fig. S1).
We tested the interaction between purified NUP153 constructs with different numbers of FG-motifs (3 μM) and capsid nanotubes assembled from purified CA protein (100 μM) in a copelleting assay (Fig. 1 and SI Appendix, Figs. S1 and S2). For this purpose, A14C/E45C disulfide cross-linked CA tubes (30) and inositol hexakisphosphate (IP6)-stabilized wild-type (WT) CA tubes (31) were employed because of their stability in low salt conditions. Consistent with previous findings (24, 26), the C-terminal tail (residues 1411 to 1475, NUP1531411–1475) efficiently bound to A14C/E45C-stabilized CA tubes. Moreover, the preceding NUP153 region (residues 896 to 1400, containing 23 FG-repeats, and 896 to 1300, with 15 FG-repeats) or a model FG-NUP, NSP1 (Saccharomyces cerevisiae orthologue of NUP62), which has 32 FG-repeats, showed very weak binding, respectively. These data indicated that FG-repeats in general do not confer efficacious binding to HIV-1 capsid in the copelleting assay and that the C terminus of NUP153 (65 aa), containing a single specific FG-motif, primarily dictated NUP153 binding to capsid (Fig. 1). These results were confirmed in copelleting assays using WT CA tubes in physiological salt (150 mM NaCl) with IP6 (1 mM) (SI Appendix, Fig. S2). These observations, together with the reported low binding affinity of the NUP153 FG-containing peptide to CA hexamers (21, 27), highlighted that FG-motifs have limited inherent capsid-binding activity. By contrast, we observed complete copelleting of NUP1531411–1475 with CA tubes even at comparatively low NUP concentrations (3 μM) (Fig. 1B and SI Appendix, Fig. S1). Since the nanotubes used here present the higher-order lattice of the assembled capsid, it suggested the existence of an additional interaction beyond the hexamer-specific NUP153 FG-motif binding site.
NUP153 Targets CA Lattice Interfaces in the Assembled Capsid.
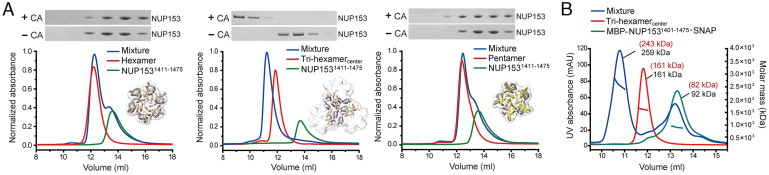
To provide a more detailed understanding of how NUP153 binds HIV-1 capsid, we analyzed the interaction between NUP1531411–1475 and various CA assemblies representing different patterns on the capsid. We used SEC to assess the migration positions of NUP1531411–1475 alone, or in the presence of individual capsomeres (CA hexamers or pentamers), as well as an assembly (termed tri-hexamercenter in this study) that mimics the interface between three hexamers (32) (Fig. 2A). In this way, a shift in migration to higher molecular weight species would provide evidence for complex formation and hence effective binding. Consistent with the reported low binding affinities (21, 27), neither CA hexamer nor pentamer detectably bound NUP1531411–1475. By contrast, efficient binding of tri-hexamercenter to NUP1531411–1475 was readily observed (Fig. 2A). We further used SEC-multiangle light scattering (SEC-MALS) to measure the molecular weight distributions in each elution peak to deduce the stoichiometric ratios of NUP153 and tri-hexamercenter in the complex. The results showed good agreement with a 1:1 molar ratio of NUP1531401–1475: tri-hexamercenter (Fig. 2B), pointing to a model where the C-terminal tail of NUP153 engages interhexamer interfaces on the capsid. These observations revealed that NUP153 preferentially engages the higher-order lattice within the assembled capsid, implying that capsid interactions with the NPC require at least a partially assembled structure.
Fig. 2.
NUP153 targets CA lattice interfaces in the assembled capsid. (A) SEC coelution assays of CA hexamer (Left), tri-hexamercenter (Middle), or pentamer (Right) with NUP1531411–1475. Top: SDS-PAGE of eluates. Only the tri-hexamercenter induced a notable peak shift of NUP1531411–1475, indicative of complex formation, with a corresponding shift of eluted NUP1531411–1475 protein by SDS-PAGE (Top). Independent experiments were repeated three times, with representative examples shown. (B) SEC-MALS analysis of NUP1531401–1475 and tri-hexamercenter. Overlay of the individual SEC-MALS elution profiles of NUP1531401–1475 (green), tri-hexamercenter (red), and a mixture of the two (blue). The measured (black) and predicted (red) molecular weights are shown.
A RRR Motif at the NUP153 C terminus Binds to the threefold Tri-Hexamer Interface in the Capsid.
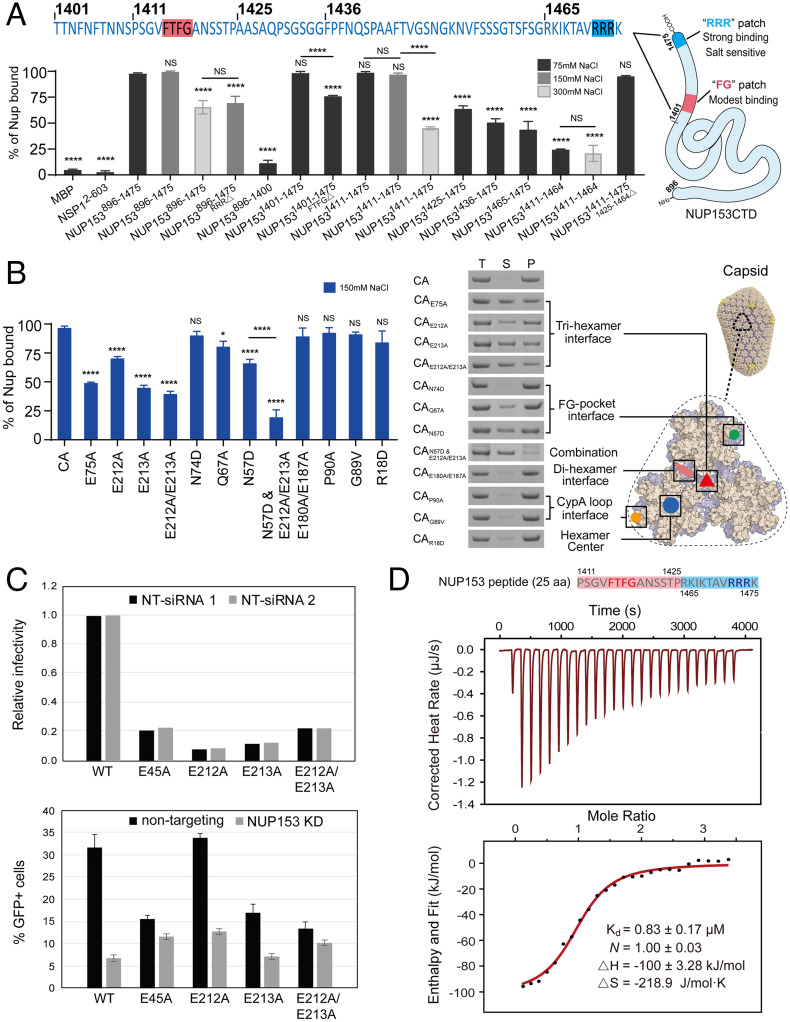
The higher-order capsid lattice preference of NUP1531411–1475 suggested that it may contain another binding site beyond the CA hexamer-targeting FG-motif. We performed a comprehensive mapping study to identify the specific region responsible for additional binding with the capsid (Fig. 3A and SI Appendix, Fig. S3). The results confirmed that the deletion or truncation of the FG-motif (NUP1531401–1475FTFG△, NUP1531425–1475, or NUP1531436–1475) reduced but did not abolish the binding (Fig. 3A and SI Appendix, Fig. S3). Interestingly, when the N-terminal region of the NUP153 C-terminal tail was progressively removed, leaving just the last 11 aa of the protein remaining without the FG-motif (NUP1531465–1475), a substantial level of CA tube binding was retained. Concomitantly, removal of the last 11 aa region (NUP1531411–1464) significantly decreased the binding. These results indicated that the C-terminal 11 residues of NUP153 may contain the additional capsid-binding sequence (Fig. 3A and SI Appendix, Fig. S3). The short 11 aa C-terminal tail of NUP153 is enriched in positively charged amino acids, including a RRR motif that resembles the capsid-binding motif in the HIV-1 restriction factor MxB (20, 33–35). Since the MxB RRR-motif targets capsid through electrostatic interactions, we tested the effect of increasing salt concentrations on CA nanotube binding. NUP153CTD bound CA tubes similarly at 150 mM NaCl and 75 mM NaCl (Fig. 3A and SI Appendix, Figs. S1 and S2). However, when the NaCl concentration was increased to 300 mM, NUP1531411–1475 lost its binding capacity significantly, while NUP1531411–1464 containing the FG-motif but not the 11-aa tail showed no change in its binding (Fig. 3A and SI Appendix, Fig. S3). This salt sensitivity suggested that the RRR-motif engaged the capsid in an MxB-like manner (36), perhaps binding at the negatively charged tri-hexamer center. We further tested CA tube copelleting of NUP153896–1475 in high salt (300 mM NaCl) alongside the NUP153896–1475 RRR-deletion mutant (Fig. 3A and SI Appendix, Fig. S3). The data showed that either high salt or loss of the RRR-motif significantly affected NUP153896–1475 binding to CA tubes, further supporting the capsid-binding property of the RRR-motif.
Fig. 3.
A bipartite interaction between NUP153 and the CA lattice. (A) Top: The C-terminal 75 aa of NUP153. Bottom: percentages of various NUP153 variants, along with maltose-binding protein (MBP) and yeast NUP NSP1 negative controls, bound to CA tubes in the copelleting assays under 75 mM NaCl (black bar), 150 mM NaCl (gray bar), or 300 mM NaCl (light gray bar) conditions. Data were expressed as mean ± SD of three independent experiments. Differences were determined by one-way ANOVA and Tukey’s multiple comparisons test, compared with NUP153896–1475; NS, not significant (P ≥ 0.05); ****P < 0.0001. The schematic of the two capsid-binding motifs on NUP153CTD is shown in the Right panel. (B) Copelleting assays of CA mutant nanotubes with NUP1531411–1475. Left: percentages of NUP1531411–1475 bound to CA mutant tubes in the copelleting assays under 150 mM NaCl. Data were expressed as mean ± SD of three independent experiments. Differences were determined by one-way ANOVA and Tukey’s multiple comparisons test, compared with A14C/E45C CA tubes, NS, not significant (P ≥ 0.05), *P < 0.05, ****P < 0.0001. Right: SDS-PAGE analysis of copelleted NUP1531411–1475 alongside the positions of mutated CA residues on the tri-hexamercenter model. Negative-stain TEM validation of the formation of various CA nanotubes is shown in SI Appendix, Fig. S4. (C) Analysis of HIV-1 infection in NUP153-depleted cells. HeLa cells were treated with two separate nontargeting control small interfering RNAs (siRNAs) or with two different NUP153-targeting siRNAs. Cell monolayers were subsequently challenged with HIV-GFP reporter viruses at dilutions appropriate for detecting infection. The fraction of infected cells was analyzed by flow cytometry for GFP expression. Top, relative infectivity of the virus stocks on control siRNA-transfected cells. Shown are the individual results from infections of cells transfected with two different nontargeting siRNAs. Bottom, levels of infection in control and NUP153-depleted cells. Shown are averages of duplicate infections with error bars designating the range of the two values obtained from the two different siRNAs. Results are representative of two independent experiments. (D) The NUP1531411–1475/1425–1464△ peptide (25 aa) binds to CA tri-hexamercenter as measured by ITC. Independent experiments were performed three times, and a representative example is shown. Curve fitting errors to a single data set are shown.
To identify the capsid surface that is targeted by the RRR-motif of NUP153, we mutated key CA residues that are located in different regions of the capsid, including the tri-hexamer interface (E212A, E213A, E212A/E213A, E75A), the di-hexamer interface (E180A/E187A), the FG-pocket (N74D, Q67A, N57D), the CypA-binding loop (P90A, G89V), and the hexamer central pore (R18D) (21, 27, 36–38). Copelleting results showed that alterations of key residues at the tri-hexamer interface (E212A, E213A, E212A/E213A, and E75A) substantially reduced NUP1531411–1475 binding to CA nanotubes (Fig. 3B and SI Appendix, Fig. S4). Mutations at the FG-pocket of CA led to significant reductions of NUP153 binding with the Q67A and N57D CA mutants, while N74D CA did not obviously impact NUP153 binding (Fig. 3B and SI Appendix, Fig. S4). The result with N74D is consistent with the previous report that the residue was key to the interaction with CPSF6, but had little effect on interaction with NUP153 (24). The combination of N57D with additional E212A/E213A mutations resulted in a nearly complete loss of NUP153 binding (Fig. 3B and SI Appendix, Fig. S4), which recapitulates the results observed for the corresponding FG and RRR mutants. By contrast, mutations in the di-hexamer interface, the CypA-binding loop, or the hexamer central pore did not significantly impact binding (Fig. 3B and SI Appendix, Fig. S4). These data are consistent with the notion that electrostatic contact between the positively charged RRR-motif and negatively charged CA tri-hexamer interface provides an extra driving force for this virus–host interaction. These data further highlight the importance of the bipartite motif of NUP153 (FG and RRR) for targeting the CA lattice beyond its hexameric capsomere building block.
To determine whether the binding of NUP153 to the tri-hexamer interface has biological consequences for infection, we depleted NUP153 in HeLa cells by siRNA transfection (SI Appendix, Fig. S5) and challenged the cells with HIV-1 mutants encoding alanine substitutions for E212 and E213. As a positive control, we included the E45A mutant, previously shown to exhibit reduced dependence on NUP153 (24, 39, 40). E45A is reported to form a hyperstable capsid (41, 42), which may interfere with the capsid property needed for stable NPC docking and the nuclear transport of the HIV-1 core. All of the mutant viruses were markedly reduced in the infection of control HeLa cells (Fig. 3C). While infection by the E212A and E213A single-mutant viruses was reduced by NUP153 depletion, though perhaps not to the extent of WT HIV-1 (Fig. 3C), infection by the E212A/E213A mutant was affected only minimally by NUP153 depletion, to an extent similar to that observed with the E45A mutant virus. These results show that the E212A/E213A double mutation in CA markedly reduces the dependence of HIV-1 infection on NUP153 in target cells, consistent with our biochemical binding results (Fig. 3B).
A 25 aa Peptide Derived from the NUP153 Bipartite Motif Strongly Binds Capsid.
To test the hypothesis that NUP153 binds the capsid in a bipartite mode using both the FG-motif and the RRR-motif, we created a construct directly connecting the FG-motif and the RRR-motif (NUP1531411–1475/1425–1464△). The resultant 25 aa segment bound CA nanotubes as efficiently as the intact CTD (NUP153896–1475), thus confirming that these two regions are sufficient to recapitulate most of the capsid-binding capacity (Fig. 3A). We further used isothermal titration calorimetry (ITC) to quantify the binding affinity of this 25 aa-peptide to the lattice-mimicking tri-hexamercenter. This minimal NUP153CTD peptide bound to tri-hexamercenter with a Kd of 0.87 ± 0.16 μM (mean and SD from three independent experiments), which is almost two orders of magnitude tighter than the very low binding affinity (Kd) reported for FG-motif peptide–hexamer binding (21, 27) (Fig. 3D).
Molecular Dynamics (MD) Simulations Identify the Tri-Hexamer Interface as the Site of NUP153 RRR Motif–Capsid Interaction.
Atomic MD simulation has been widely used to study the dynamics of HIV-1 capsid and its interactions with cellular cofactors and drug-like molecules (36, 43–47). To investigate the NUP153CTD-CA interactions at the tri-hexamer interface, a NUP1531451–1475–CA tri-hexamercenter complex was built (SI Appendix, Fig. S6A) and simulated for 8.7 μs. In the simulation, NUP1531451–1475 was found to be mostly unstructured at the CA tri-hexamer interface. The secondary structure calculation (SI Appendix, Fig. S6B) of NUP1531451–1475 from the simulation indicated that it is highly disordered. The NUP1531451–1475 peptide folded into transient turns and short helices, until a relatively stable secondary structure of a short turn centering around residues 1467 to 1470 was formed at about 8 μs. Markov state model analysis (48–50) indicated that the NUP1531451–1475 peptide was highly dynamic, identifying several metastable states of NUP1531451–1475 at the tri-hexamer interface (SI Appendix, Figs. S6C and S7). Among these metastable states, the state 8 was relatively stable, corresponding to the fold appearing at around 8 μs, while NUP1531451–1475 rapidly transited between other metastable states in the rest of the simulation. The RRR motif of NUP1531451–1475 inserted deeply into the tri-hexamer interface in state 8, forming extensive interactions with the negatively charged glutamate residues from four different CA monomers (SI Appendix, Fig. S6D and Table S1). Because of the relative conformational stability of NUP1531451–1475 and the extensive interactions with CA, the state 8 was considered as the binding state of the NUP153CTD at the CA tri-hexamer interface, although the overall interaction appeared to be transient and not dominated by a single stable state. The occupancies of the top NUP153 RRR-motif residues in contact with CA tri-hexamercenter were mostly in the range of 30 to 40% throughout the entire simulation (SI Appendix, Table S2), in contrast to the 60 to 95% occupancies previously observed in the MD simulation with the MxB RRR-motif (36). This difference reflects the effect of the RRR flanking sequence, which is very different between MxB and NUP153, in HIV-1 capsid interactions. The dynamic character of the capsid–NUP153 RRR-motif interaction plausibly evolved to facilitate the transient nature of the nuclear import process.
Cellular NUP153 Associates with HIV-1 Capsid Assemblies.
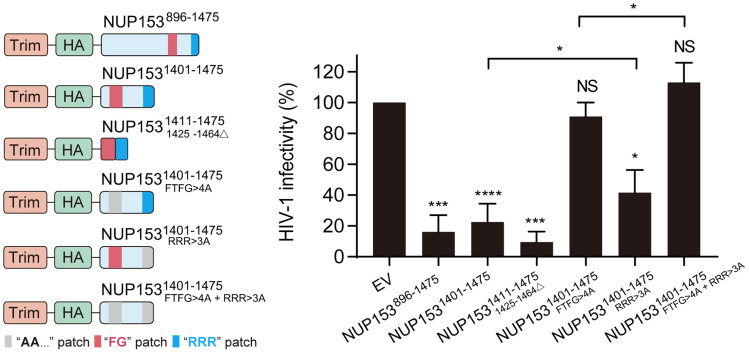
To validate the results of the in vitro binding assays under HIV-1 infection conditions, we employed the previously described TRIM-NUP153CTD construct wherein CTD residues 896 to 1475 were fused to the RING, B-box 2, and coiled-coil domains of rhesus TRIM5α (24). Effector functions provided by the TRIM domains coupled with capsid binding, which was provided by the NUP153CTD, significantly inhibited HIV-1 infection (24) (Fig. 4). Deletion mutagenesis revealed that TRIM-NUP1531401-1475 lacking the 505 N-terminal residues of 896-1475, as well as a minimal fusion construct that harbored just the 25 residues composed of the FG-motif (1411 to 1424) and the RRR-motif (1465 to 1475), conferred full restriction activity (Fig. 4). These results revealed that the minimal NUP153 bipartite motif fragment was associated with HIV-1 capsid during virus infection. To determine the individual roles of the FG- versus RRR-motifs in HIV-1 core binding, we constructed TRIM-NUP1531401–1475 variants with single-motif mutations (FTFG>4A or RRR>3A) as well as the combined FTFG>4A + RRR>3A double-motif mutations. The FTFG>4A mutant significantly reduced the ability of TRIM-NUP153 to inhibit HIV-1 infection, while the RRR>3A mutant slightly reduced the inhibitory activity. Consistent with this observation, the addition of RRR>3A to the FTFG>4A construct slightly improved HIV-1 infection (Fig. 4). A number of parameters could have contributed to the apparent modest effect of the RRR>3A mutation in the TRIM-fusion assays as compared to its effect in biochemical binding assays (Fig. 3A). The physiological function of NUP153, as part of the NPC, in HIV-1 infection is to aid the nuclear import of the viral core. By contrast, TRIM-mediated restriction occurs in the cell cytoplasm (24).
Fig. 4.
Contributions of NUP153 FG and RRR motifs to capsid binding in cells. Restriction of HIV-1 infection by TRIM-NUP153 fusion proteins. Schematic of test constructs (Left) and results (mean ± SD of three independent experiments) of HIV-1 infection assays (Right) are shown. EV, empty vector. Statistical significance was determined by two-tailed Student’s t test; NS, not significant (P ≥ 0.05); *P < 0.05; ***P <0.05; <0.001; ****P < 0.0001. Immunoblot of protein expression is shown in SI Appendix, Fig. S8A. HA, hemagglutinin tag.
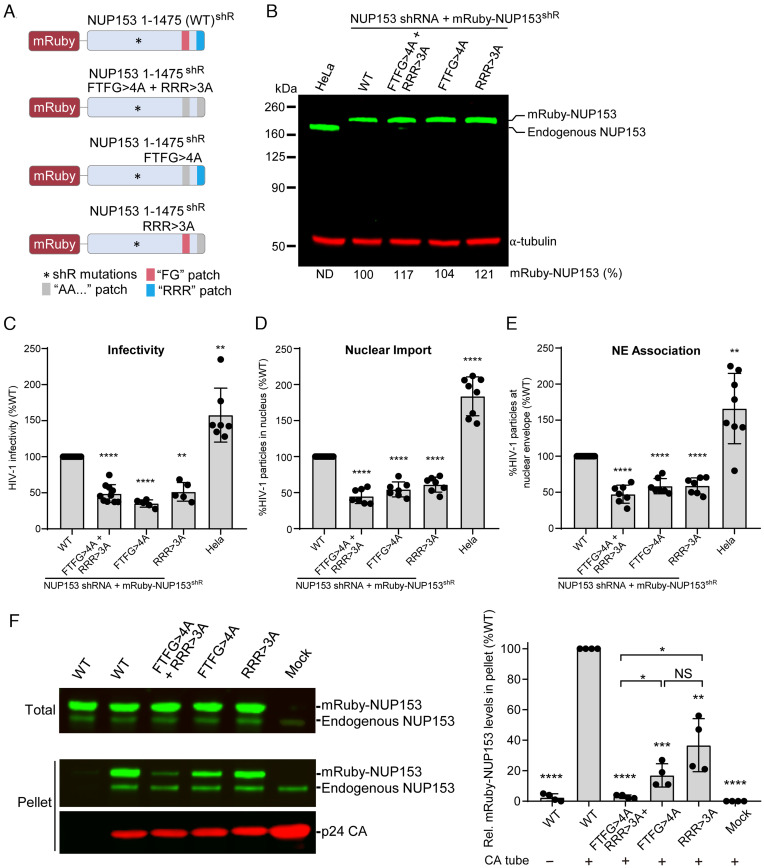
Both the FG-motif and the RRR-motif Are Important for the Nuclear Import of HIV-1 Cores.
To further determine the role of the NUP153 FG- and RRR-motifs in HIV-1 nuclear import, HeLa cell lines were generated by stable knockdown of endogenous NUP153 with short hairpin RNA (shRNA) and stable expression of shRNA-resistant, mRuby-tagged WT, double-mutant (FTFG>4A + RRR>3A), or single-mutant (FTFG>4A or RRR>3A) NUP153 (Fig. 5A). Immunoblotting of cell lysates for NUP153 indicated that endogenous NUP153 was undetectable upon shRNA-mediated knockdown and that the expression of the shRNA-resistant WT and mutant mRuby-NUP153 was similar to or higher than the level of endogenous NUP153 in the parental HeLa cells (Fig. 5B). WT and mutant mRuby-NUP153 proteins primarily localized to the nuclear envelope (SI Appendix, Fig. S9A). The growth kinetics of pools of transduced cells expressing the mRuby-NUP153 WT or mutants were similar to the parental HeLa cells, indicating that the expression of the exogenous NUP153 proteins restored NUP153 function with no apparent cytotoxicity (SI Appendix, Fig. S9B). To determine the role of the NUP153 FG- and RRR-motifs in HIV-1 infectivity, the mRuby-NUP153 expressing cells were infected with a single-cycle HIV-1 GFP reporter virus and the percentage of GFP-expressing cells was determined ~30 h after infection. A twofold decrease in infectivity was observed in cells expressing the double-mutant (FTFG>4A + RRR>3A) or single-mutant (FTFG>4A or RRR>3A) NUP153 compared to cells expressing WT NUP153 (Fig. 5C).
Fig. 5.
Important roles for NUP153 FG- and RRR-motifs in nuclear import of HIV-1 cores. (A) Schematic of mRuby-NUP153 expression constructs. HeLa cell lines were generated by knockdown of endogenous NUP153 with shRNA and expression of shRNA-resistant (shR) mRuby-NUP153 containing either WT NUP153, or NUP153 mutants FTFG>4A + RRR>3A, FTFG>4A only, or RRR>3A only. (B) Immunoblot of mRuby-NUP153 and endogenous NUP153 protein expression. Expression levels were quantified and are shown as a percentage of mRuby-NUP153 in HeLa cells. ND, not detected. α-tubulin was used as a loading control. (C) HIV-1 infection assay. Results are mean ± SD of ≥5 independent experiments. (D and E) The percentage of total (cytoplasmic + nuclear) GFP content marker-labeled HIV-1 cores inside the nucleus (D) and the percentage of cytoplasmic GFP-labeled HIV-1 cores at the nuclear envelope (NE) (E) were determined at 6 h postinfection. Results are mean ± SD of ≥5 independent experiments. Statistical significance was determined by two-tailed Welch’s t test; **P<0.01; ****P < 0.0001. (F) Left: copelleting assays of CA nanotubes with full-length mRuby-NUP153 and mutants from HEK293T cell lysates. Endogenous NUP153 served as an internal control for binding. Right: percentages of NUP153 mutants bound to CA tubes in the copelleting assays (normalize by WT). Data were expressed as mean ± SD of four independent experiments. Statistical significance was determined by two-tailed Welch’s t test; NS, not significant (P ≥ 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Relative mRuby-NUP153 and mutant expression levels are shown in SI Appendix, Fig. S8B.
To determine the role of the NUP153 FG- and RRR-motifs in nuclear import, virions labeled with GFP content marker (10) were used to infect the different mRuby-NUP153 cell lines. The infected cells were fixed 6 h postinfection and the nuclear DNA was stained using DAPI. Confocal images of the infected cells were acquired and the localization of the GFP-labeled HIV-1 cores in the cytoplasm, at the nuclear envelope (i.e., edge of DAPI), or inside the nucleus, was determined. The percentage of total (cytoplasmic + nuclear) GFP-labeled HIV-1 cores inside the nucleus was ~twofold lower in the NUP153 mutant-expressing cell lines compared to the NUP153 WT-expressing cell line, indicating that both the FG- and RRR-motifs are similarly important for the nuclear import of HIV-1 cores (Fig. 5D). The decrease in nuclear import efficiency correlated with the decrease in infectivity, indicating that the reduced infectivity was primarily due to the reduced nuclear import. The percentage of cytoplasmic HIV-1 cores that colocalized with the nuclear envelope was also reduced ~twofold in cells expressing the mutant NUP153 constructs compared to the level in cells expressing WT NUP153 (Fig. 5E), indicating that the decrease in nuclear import was likely due to reduced stable docking of the HIV-1 cores with the NPCs that contain the NUP153 mutant proteins. These observations suggest that HIV-1 core interactions with NUP153 FG- and RRR-motifs are critical for stable viral core docking with the nuclear envelope. These results validate that NUP153 facilitates HIV-1 core docking and nuclear entry via two binding regions: The first is the previously reported FG-hexamer interface; the second is the positively charged RRR-motif targeting the CA tri-hexamer interface (Fig. 3).
To further address the roles of the FG versus RRR motifs in capsid binding under physiological conditions, we performed CA tube copelleting assays with full-length cellular NUP153 or its missense mutant forms. HEK293T cells were transfected with WT or mutant mRuby-NUP153 expression plasmids and the resulting cell lysates were used for copelleting with disulfide cross-linked 14C/45C CA tubes. The results showed that full-length endogenous NUP153 (internal control) and mRuby-NUP153 copelleted efficiently with CA tubes, while mutation of either FTFG>4A or RRR>3A significantly reduced copelleting, and double mutation of the FTFG- and RRR-motifs reduced binding to background levels (Fig. 5F). These data are consistent with the biochemical binding results using purified NUP153 constructs (Fig. 3) and support the notion that the NUP153 bipartite FG and RRR-motif is important for interacting with HIV-1 capsid.
NUP153 Stabilizes Tubular Capsid Assemblies through the Bipartite Motif Interaction.
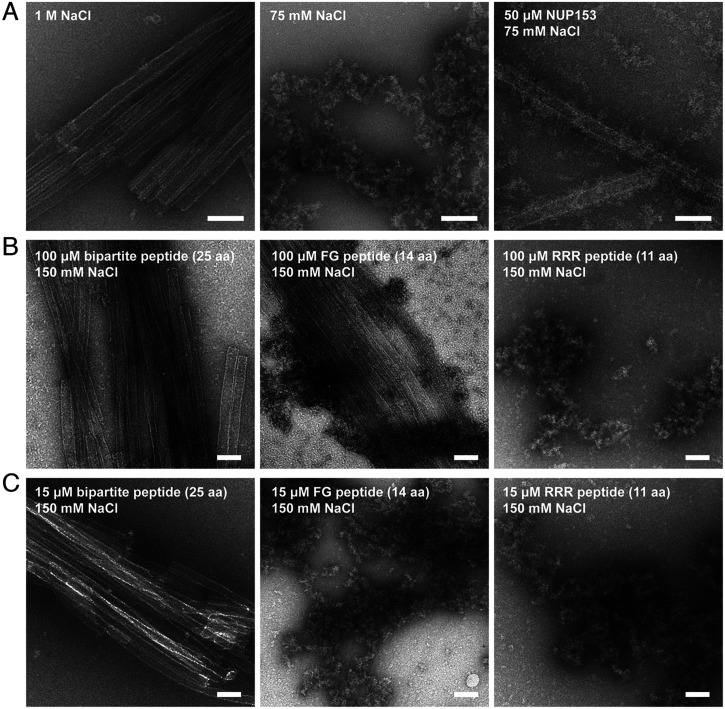
How and when the capsid uncoats during HIV-1 infection remains a longstanding question (51). To better understand the potential effect of NUP153 in HIV-1 uncoating during nuclear import, we tested its effect on WT CA tubes under different salt conditions. The WT CA tube was stable in a high salt environment such as 1 M NaCl, but in the absence of IP6, its lattice structure rapidly collapsed and disintegrated when the salt concentration was lowered to 75 mM (Fig. 6A). However, tube disassembly at low salt concentration without IP6 was averted in the presence of NUP1531411–1475, resulting in a large number of intact tubes coated with fuzzy, irregularly shaped NUP1531411–1475 proteins (Fig. 6A). These results suggested that the CA lattice was stabilized by NUP153 binding. The stabilizing effect of NUP1531411–1475 was concentration dependent (SI Appendix, Fig. S10). A large number of micrometer-long WT CA tubes (6 µM CA) were observed at 50 μM NUP153, some tubes of submicrometer in length remained at 5 μM NUP153, and a complete loss of tube structures was observed at 1 μM NUP153 (SI Appendix, Fig. S10). These results suggest that, given its high local concentration in the NPC, NUP153 binding to the capsid likely does not trigger uncoating but rather helps stabilize the CA lattice as the capsid passes through the central channel.
Fig. 6.
NUP153 stabilizes CA assemblies. (A) Negative-stain electron micrographs of intact WT CA tubes under high salt conditions (1 M NaCl, Left), disintegrated WT CA tubes under low salt conditions (75 mM NaCl, Middle), and WT CA tubes (6 µM CA) stabilized by NUP1531411–1475 (50 µM) under low salt conditions (75 mM NaCl, Right). (Scale bars: 100 nm.) (B) WT CA tubes (12 µM CA) incubated with 100 µM bipartite peptide (25 aa), FG-motif peptide (14 aa) and RRR-motif peptide (11 aa) under 150 mM NaCl. (Scale bars: 100 nm.) (C) Same assays as in (B), but with the peptides at 15 µM with WT CA tubes. (Scale bars: 100 nm.) All experiments were performed independently three times, with representative examples shown.
We next tested various NUP153 constructs to determine the contributions of the FG- and RRR-motifs to CA lattice stabilization. The 25 aa-peptide (NUP1531411–1475/1425–1464△,15 μM) containing both motifs efficiently stabilized WT CA tubes (12 µM CA) at 150 mM NaCl (Fig. 6 B and C). We further synthesized the FG-motif only NUP1531411–1424 peptide (14 aa) and the RRR-motif only NUP1531465–1475 peptide (11 aa). The FG-motif peptide at a high concentration (100 μM) could stabilize WT CA tubes under 150 mM salt (Fig. 6B), but could not maintain CA tube stability when the peptide concentration was reduced to 15 μM (Fig. 6C). By contrast, the RRR-motif peptide (11 aa) failed to stabilize WT CA tubes (Fig. 6 B and C). The results suggest that the FG-motif provides most of the direct stabilizing ability, potentially with a mechanism similar to that of the capsid-tabilizing Gilead compound GS-6207 which also targets the FG-binding pocket of CA, albeit the stabilization by the NUP153 FG-motif is limited due to its weak binding to the CA hexamer (52). However, when both FG- and RRR-motifs are present, the RRR-motif enhances binding to the CA lattice, allowing lower concentrations of the bipartite peptide to efficiently stabilize CA assemblies (Fig. 6C). These results support the notion that both the FG- and RRR-regions in the NUP153 bipartite motif are needed to efficiently stabilize the capsid, although it should be noted that stabilization effects on CA tubes may be different from that on the cone-shaped HIV-1 capsid, as previously reported for other capsid-stabilizing compounds and cofactors (27, 52–57).
Discussion
HIV-1 nuclear import is one of the least understood processes of the viral life cycle (58). It was initially thought that the capsid immediately uncoated upon viral entry into the cell, with only recent studies pointing to the possibility of a largely intact capsid passing through the NPC into the nucleus (8–10). However, mechanistic data regarding the transport of HIV-1 cores through the NPC remain scarce. This study provided in-depth mechanistic insights into how the HIV-1 capsid interacts with a critical NPC component, NUP153, pointing to a likely important step of the import mechanism. We showed that the 65 aa C-terminal tail of NUP153, containing a single FG-motif and an RRR-motif, provides most of the capsid-binding capacity of the extended (aa 896 to 1475) CTD. The canonical FG-motif targeted the CA hexamer building block. Beyond that, a previously unexplored RRR-motif at the NUP153 C terminus interacted with HIV-1 capsid at the interface of tri-hexamers. HIV-1 infection studies indicated that the bipartite motif of NUP153 interacted with the capsid, and both FG- and RRR-motifs were important for the infectivity and the nuclear import of HIV-1 cores. We also created a minimal 25 aa peptide comprising the FG- and RRR-motifs that retained potent capsid binding in vitro and in the context of HIV-1 infection (Figs. 3D and 4A). As the HIV-1 capsid is an attractive target for therapeutic intervention (52, 57, 59, 60), this efficient capsid-targeting peptide may represent a starting point for development of a unique HIV-1 inhibitor targeting nuclear import.
Notably, the most C-terminal FG-repeat of NUP153 conferred substantial CA binding affinity (Fig. 1B), which is consistent with previous studies (24, 26). The preceding NUP153 region, which contains 23 additional FG-repeats, imparted very weak interaction with CA tubes by themselves and only moderately contributed to binding in the presence of the NUP153 C-terminal tail (Fig. 3A). This observation indicates that the FG di-residue alone is not sufficient for stable engagement of HIV-1 capsid, and that flanking residues also play important roles. We performed bioinformatic analysis to identify the determinants that enable interaction with the capsid. However, no significant features in the immediate flanking sequence of the FG-motifs could be determined. It is possible that capsid interaction-enabling flanking residues function in unique ways without a common mechanism, either through direct contacts or facilitating restructuring of the FG di-residue for better interactions. In addition, it is known that FG-NUPs form a selective diffusion barrier governing the transport of macromolecules through the NPC (14, 15). As FG-motifs affect the intrinsic properties of NUPs, such as the cohesiveness that impacts the state and function of the NPC central channel (61, 62), the other FG-repeats on NUP153 may indirectly influence HIV-1 nuclear transport through their NPC channel-conforming properties. A more complete understanding of the mechanisms dictating the selective interactions between HIV-1 capsid and the FG-motifs in NUP153 and other NUPs must await future studies.
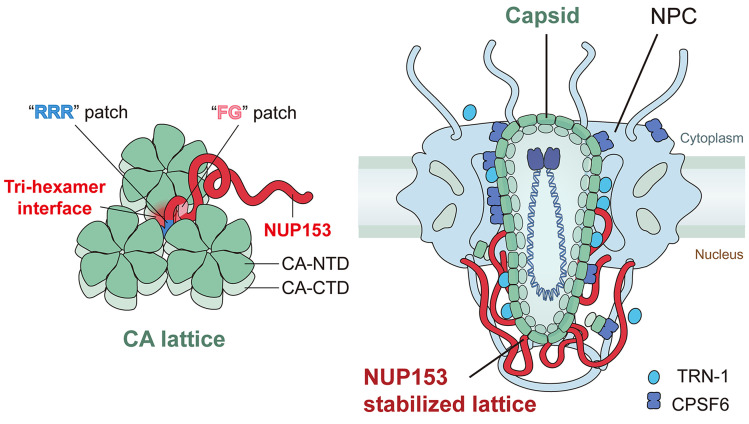
The NUP153 C-terminal region recognizes a pattern at the tri-hexamer center that only exists in the assembled capsid lattice. This binding characteristic has profound implications on the mode by which the capsid engages the NPC to enter the nucleus. Although there is evidence that NUP153 may be sufficiently unstructured to extend into the cytosol under some circumstances (25), it is anchored at the NPC nuclear basket and thus positioned in the nucleoplasm. The higher-order CA lattice targeting mode of NUP153 supports that nuclear penetration of the capsid at least needs to be in a partially assembled form, especially if a majority of the 32 copies of NPC-associated NUP153 engage the capsid during nuclear import (Fig. 7). These findings are most consistent with a recent model where the capsid passes through the NPC in an intact or largely intact form (8–10).
Fig. 7.
Schematic of how the CA lattice engages NUP153 to mediate HIV-1 capsid nuclear entry.
Our data demonstrated the functions of the bipartite NUP153 C-terminal FG- and RRR-motifs in HIV-1 capsid binding and nuclear import. Both FG- and RRR-motifs from purified or cellular NUP153 proteins exhibited robust interactions with CA assemblies and were important for the nuclear import of HIV-1. Interestingly, simultaneous mutations of both motifs did not reveal an additive effect on HIV-1 nuclear import, which may hint that both motifs are needed, potentially in a cooperative manner, and that mutant proteins lacking either motif would “switch off” this particular NUP153 function. We cannot rule out the possibility that the RRR-motif works in an indirect way, but still functionally couples with the FG-motif to influence HIV-1 nuclear import. It should be noted that the RRR-motif displayed a weak effect in the TRIM-fusion assay that detects capsid engagement in the cytoplasm. This is potentially due to the many capsid-binding factors (17, 36, 38, 63–67) in the cytoplasmic environment that may be absent from the physiological environment of NUP153, given its localization on the nuclear face of NPC. It is also possible that a nonlinear relationship exists between capsid-binding and TRIM-mediated HIV-1 restriction. Since many other factors are simultaneously involved in HIV-1 nuclear import (58, 68–71), and NPCs are known to be pleiomorphic and dynamic (20, 72–74), more extensive work is needed to fully delineate the roles of the two NUP153 motifs in the context of HIV-1 infection, especially in relevant primary cell types.
Our results are consistent with the notion that NUP153 binding helps stabilize the capsid lattice as it passes through the central channel of the NPC. Recent studies have indicated that the capsid could remain at the nuclear pore for a few hours during its transit through the NPC (8, 75). NUP153’s stabilization effect may play a significant role in keeping the capsid intact or largely intact throughout the transport process. More studies are needed to validate and probe the extent of the stabilization of HIV-1 capsid, as the stabilizing effect was inferred from our assays done with CA tubular assemblies. In addition to NUPs, other proteins, such as CPSF6 and transportin 1 (TRN-1), interact with the HIV-1 capsid to facilitate its nuclear import (67, 76, 77). These proteins may modulate capsid stability in opposite ways to achieve a fine balance to allow its passage through the NPC. It has been reported that CPSF6 (78, 79) and TRN-1 (67) deform or disrupt CA tubes. In addition, it has been reported that reverse transcription completes inside the assembled capsid after nuclear entry and the progress of reverse transcription may have dramatic effects on the biophysical properties of HIV-1 cores (80, 81). The combined effect of these factors may enable remodeling of the capsid at the NPC central channel such that an intact or largely intact capsid enters into the nucleus (Fig. 7). It has been observed that the capsid remains cone- or tube-shaped inside the nucleus, however, with a modified CA lattice after passing through the NPC (9). Taken together, one can picture a scenario whereby multiple copies of NUP153 at the NPC basket bind and position the capsid (82), protect the integrity of the CA lattice, while together with other cellular factors help remodel the capsid to enable its nuclear import (Fig. 7). Future experiments are needed to reveal whether CA lattice remodeling indeed occurs, and, if so, the details of the remodeling and how this process is regulated at the NPC.
Materials and Methods
Experimental details and methods can be found in SI Appendix, including sources of proteins, plasmids, procedures for protein purification, and descriptions of the preparation of CA assemblies used in the experiments. Methods to investigate NUP153–CA interactions, including CA tube copelleting assays, SEC coelution assays, isothermal titration calorimetry, and SEC linked to multiangle light scattering (SEC-MALS) are described in SI Appendix. Details of MD simulations of NUP153 RRR motif–capsid interaction are described in SI Appendix, including the procedures of the simulation, Markov state model analysis and validation. For the virology assays, including the TRIM-NUP153-mediated restriction assays, HIV-1 infection assays in NUP153-depleted cells, and the live-cell imaging of virus nuclear entry experiments, all sources of cell lines, procedures for their maintenance, plasmid constructions, procedures of experiments, and data analysis are described in SI Appendix. The details of negative-stain electron microscopy imaging and statistical analysis are also available in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dynamics of NUP1531451-1475 at CA tri-hexamer interface from MD simulation. The NUP153 peptide is shown as a red ribbon, while the HIV-1 CA N-terminal domain (NTD) and CTD are shown as golden and blue ribbons, respectively. NUP1531451-1475 was found to be mostly unstructured during the simulations, except for a short α-helix at the C-terminus (residue 1470 to 1475) that appeared temporarily.
Acknowledgments
We thank the Xiong Lab for discussions. This work was supported in part by NIH grants P50AI150481 (Y.X., C.A., and A.N.E.), U54AI170791 (Y.X., C.A., and A.N.E.), R01AI162260 (Y.X., C.L., and C.P.L.), P30GM110758 (J.R.P.), R01AI052014 (A.N.E.), R21GM109466 (C.L. and C.P.L.), and R01GM105672 (C.P.L.); the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by Intramural AIDS Targeted Antiviral Program grant funding (V.K.P.); Collaboration Development Pilot Program awards from the Pittsburgh Center for HIV Protein Interactions (J.R.P. and C.L.); and a Singapore Agency for Science, Technology and Research Graduate Scholarship (Q.X.). Anton 2 computer time was provided by the Pittsburgh Supercomputing Center (PSC) through NIH Grant R01GM116961. The Anton 2 machine at PSC was generously made available by D. E. Shaw Research.
Author contributions
Q.S. and Y.X. designed research; Q.S., S.K., C.X., S.J., J.S., R.C.B., C.W., S.C.D., T.N.T., Y.H., S.Y., J.T., Q.F., C.A., A.N.E., J.R.P., and V.K.P. performed research; Q.S., S.K., C.X., S.J., R.C.B., L.L., Q.X., C.W., S.C.D., T.T., C.A., A.N.E., J.R.P., V.K.P., C.L., and Y.X. analyzed data; and Q.S., S.K., R.C.B., Q.X., T.T., C.P.L., C.A., A.N.E., J.R.P., V.K.P., C.L., and Y.X. wrote the paper.
Competing interests
The authors have organizational affiliations to disclose, A.N.E. has consulted for ViiV Healthcare, Co., on work unrelated to this project.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Lusic M., Siliciano R. F., Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 15, 69–82 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hulme A. E., Perez O., Hope T. J., Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 9975–9980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamashita M., Engelman A. N., Capsid-dependent host factors in HIV-1 infection. Trends Microbiol. 25, 741–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganser B. K., Li S., Klishko V. Y., Finch J. T., Sundquist W. I., Assembly and analysis of conical models for the HIV-1 core. Science 283, 80–83 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Li S., Hill C. P., Sundquist W. I., Finch J. T., Image reconstructions of helical assemblies of the HIV-1CA protein. Nature 407, 409–413 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Briggs J. A. G., Wilk T., Welker R., Krausslich H. G., Fuller S. D., Structural organization of authentic, mature HIV-1 virions and cores. Embo J. 22, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Appen A., et al. , In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdick R. C., et al. , HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. U.S.A. 117, 5486–5493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zila V., et al. , Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046.e1018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C., Burdick R. C., Nagashima K., Hu W. S., Pathak V. K., HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 118, e2019467118. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin D. H., Hoelz A., The structure of the nuclear pore complex (An update). Annu. Rev. Biochem. 88, 725–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook A., Bono F., Jinek M., Conti E., Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Beck M., Hurt E., The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Bio. 18, 73–89 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F., Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Timney B. L., et al. , Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dharan A., Bachmann N., Talley S., Zwikelmaier V., Campbell E. M., Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 5, 1088–1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bichel K., et al. , HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology 10, 81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Nunzio F., et al. , Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 440, 8–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matreyek K. A., Engelman A., The requirement for nucleoporin NUP153 during human immunodeficiency virus Type 1 infection is determined by the viral capsid. J. Virol. 85, 7818–7827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane M., et al. , Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. Elife 7, e35738. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price A. J., et al. , Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. Plos Pathog. 10, e1004459. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walther T. C., et al. , The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. Embo J. 20, 5703–5714 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz A., et al. , Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. Plos Pathog. 6, e1000741. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matreyek K. A., Yucel S. S., Li X., Engelman A., Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. Plos Pathog. 9, e1003693. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahrenkrog B., et al. , Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140, 254–267 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Buffone C., et al. , Nup153 onlocks the nuclear pore complex for HIV-1 nuclear translocation in nondividing cells. J. Virol. 92, e00648–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya A., et al. , Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. U.S.A. 111, 18625–18630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price A. J., et al. , CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. Plos Pathog. 8, e1002896. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebensburg S. V., et al. , Sec24C is an HIV-1 host dependency factor crucial for virus replication. Nat. Microbiol. 6, 435–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selyutina A., Bulnes-Ramos A., Diaz-Griffero F., Binding of host factors to stabilized HIV-1 capsid tubes. Virology 523, 1–5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallery D. L., et al. , A stable immature lattice packages IP6 for HIV capsid maturation. Sci. Adv. 7, eabe4716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summers B. J., et al. , Modular HIV-1 capsid assemblies reveal diverse host-capsid recognition mechanisms. Cell Host Microbe. 26, 203–216.e206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King M. C., Raposo G., Lemmon M. A., Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U.S.A. 101, 8957–8962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busnadiego I., et al. , Host and viral determinants of Mx2 antiretroviral activity. J. Virol. 88, 7738–7752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kane M., et al. , MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smaga S. S., et al. , MxB restricts HIV-1 by targeting the tri-hexamer interface of the viral capsid. Structure 27, 1234–1245.e1235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dick R. A., Mallery D. L., Vogt V. M., James L. C., IP6 regulation of HIV capsid assembly, stability, and uncoating. Viruses-Basel 10, 640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaller T., et al. , HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. Plos Pathog 7, e1002439. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J., Price A. J., Halambage U. D., James L. C., Aiken C., HIV-1 resistance to the capsid-targeting inhibitor PF74 results in altered dependence on host factors required for virus nuclear entry. J. Virol. 89, 9068–9079 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee K., et al. , Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang R. F., et al. , Second-site suppressors of HIV-1 capsid mutations: Restoration of intracellular activities without correction of intrinsic capsid stability defects. Retrovirology 9, 30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C., Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76, 5667–5677 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang P. J., et al. , Cyclophilin a stabilizes the mature HIV-1 capsid through a novel non-canonical binding site. Biophys. J. 108, 190a (2015). [Google Scholar]
- 44.Perilla J. R., Hadden J. A., Goh B. C., Mayne C. G., Schulten K., All-atom molecular dynamics of virus capsids as drug targets. J. Phys. Chem. Lett. 7, 1836–1844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perilla J. R., Schulten K., Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat. Commun. 8, 15959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang P. T., et al. , FEZ1 is recruited to a conserved cofactor site on capsid to promote HIV-1 trafficking. Cell Rep. 28, 2373–2385 e2377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dick R. A., et al. , Inositol phosphates are assembly co-factors for HIV-1. Nature 560, 509–512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherer M. K., et al. , PyEMMA 2: A software package for estimation, validation, and analysis of markov models. J. Chem. Theory Comput. 11, 5525–5542 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Husic B. E., Pande V. S., Markov state models: From an art to a science. J. Am. Chem. Soc. 140, 2386–2396 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wang W., Cao S., Zhu L., Huang X., Constructing Markov State Models to elucidate the functional conformational changes of complex biomolecules. WIREs Comput. Mol. Sci. 8, e1343. (2018). [Google Scholar]
- 51.Hofer U., Entering and breaking for HIV? Nat. Rev. Microbiol. 18, 264 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Bester S. M., et al. , Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 370, 360–364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faysal K. M. R., Pharmacologic hyperstabilisation of the HIV-1 capsid lattice induces capsid failure. bioRxiv, 10.1101/2022.09.21.508807 (2022). [DOI] [PMC free article] [PubMed]
- 54.Shi J., Zhou J., Shah V. B., Aiken C., Whitby K., Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dostalkova A., et al. , PF74 and its novel derivatives stabilize hexameric lattice of HIV-1 mature-like particles. Molecules 25, 1895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bester S. M., et al. , Structural and mechanistic bases of viral resistance to HIV-1 capsid inhibitor lenacapavir. Mbio 13, e0180422. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Link J. O., et al. , Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 584, 614–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen Q., Wu C. X., Freniere C., Tripler T. N., Xiong Y., Nuclear import of HIV-1. Viruses-Basel 13, 2242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thenin-Houssier S., Valente S. T., HIV-1 capsid inhibitors as antiretroviral agents. Curr. Hiv. Res. 14, 270–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carnes S. K., Sheehan J. H., Aiken C., Inhibitors of the HIV-1 capsid, a target of opportunity. Curr. Opin. Hiv Aids 13, 359–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen Q., et al. , DNA-origami nanotrap for studying the selective barriers formed by phenylalanine-glycine-rich nucleoporins. J. Am. Chem. Soc. 143, 12294–12303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher P. D. E., et al. , A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. Acs Nano 12, 1508–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin D. H., Zimmermann S., Stuwe T., Stuwe E., Hoelz A., Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J. Mol. Biol. 425, 1318–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miles R. J., et al. , MxB sensitivity of HIV-1 is determined by a highly variable and dynamic capsid surface. Elife 9, e56910. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fribourgh J. L., et al. , Structural insight into HIV-1 restriction by MxB. Cell Host & Microbe. 16, 627–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fricke T., et al. , MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernandez J., et al. , Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat Microbiol 4, 1840–1850 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Rossi E., Meuser M. E., Cunanan C. J., Cocklin S., Structure, function, and interactions of the HIV-1 capsid protein. Life-Basel 11, 100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dharan A., Campbell E. M., Teaching old dogmas new tricks: Recent insights into the nuclear import of HIV-1. Curr. Opin. Virol. 53, 101203 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bedwell G. J., Engelman A. N., Factors that mold the nuclear landscape of HIV-1 integration. Nucleic Acids Res. 49, 621–635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Temple J., Tripler T. N., Shen Q., Xiong Y., A snapshot of HIV-1 capsid-host interactions. Curr. Res. Struct. Biol. 2, 222–228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmerli C. E., et al. , Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Bley C. J., et al. , Architecture of the cytoplasmic face of the nuclear pore. Science 376, eabm9129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosalaganti S., et al. , AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376, eabm9506 (2022). [DOI] [PubMed] [Google Scholar]
- 75.Zuliani-Alvarez L., Towers G. J., Identifying a nuclear passport for HIV. Elife 8, e45580. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chin C. R., et al. , Direct visualization of HIV-1 replication intermediates shows that capsid and CPSF6 modulate HIV-1 intra-nuclear invasion and integration. Cell Rep. 13, 1717–1731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bejarano D. A., et al. , HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. Elife 8, e41800. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ning J., et al. , Truncated CPSF6 forms higher-order complexes that bind and disrupt HIV-1 capsid. J. Virol. 92, e00368–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhong Z., et al. , Cytoplasmic CPSF6 regulates HIV-1 capsid trafficking and infection in a cyclophilin A-dependent manner. mBio 12, e03142–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rankovic S., Varadarajan J., Ramalho R., Aiken C., Rousso I., Reverse transcription mechanically initiates HIV-1 capsid disassembly. J. Virol. 91, e00289–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rankovic S., Deshpande A., Harel S., Aiken C., Rousso I., HIV-1 uncoating occurs via a series of rapid biomechanical changes in the core related to individual stages of reverse transcription. J. Virol. 95, e00166–e001621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen Q., et al. , Modeling HIV-1 nuclear entry with nucleoporin-gated DNA-origami channels. Nat. Struct. Mol. Biol. (2023), 10.1038/s41594-023-00925-9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dynamics of NUP1531451-1475 at CA tri-hexamer interface from MD simulation. The NUP153 peptide is shown as a red ribbon, while the HIV-1 CA N-terminal domain (NTD) and CTD are shown as golden and blue ribbons, respectively. NUP1531451-1475 was found to be mostly unstructured during the simulations, except for a short α-helix at the C-terminus (residue 1470 to 1475) that appeared temporarily.
Data Availability Statement
All study data are included in the article and/or SI Appendix.