Biochemistry Correction for “The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties,” by Clarisse Roblin, Steve Chiumento, Olivier Bornet, Matthieu Nouailler, Christina S. Müller, Katy Jeannot, Christian Basset, Sylvie Kieffer-Jaquinod, Yohann Couté, Stéphane Torelli, Laurent Le Pape, Volker Schünemann, Hamza Olleik, Bruno De La Villeon, Philippe Sockeel, Eric Di Pasquale, Cendrine Nicoletti, Nicolas Vidal, Leonora Poljak, Olga Iranzo, Thierry Giardina, Michel Fons, Estelle Devillard, Patrice Polard, Marc Maresca, Josette Perrier, Mohamed Atta, Françoise Guerlesquin, Mickael Lafond, and Victor Duarte, which was first published July 27, 2020; 10.1073/pnas.2004045117 (Proc. Natl. Acad. Sci. U.S.A. 117, 19168–19177).
The authors note that on page 19168, left column, in the Abstract, line 8, “Here we report its unique comact structure on the basis of four intramolecular thioether bridges with reversed stereochemistry introduced posttranslationally by a specific radical-SAM sactisynthase.” should instead appear as “Here, we report its unique compact structure on the basis of four intramolecular thioether bridges introduced post-translationally by a specific radical-SAM sactisynthase.” The online version has been corrected.
The authors note that on page 19170, left column, second full paragraph, line 30, “The structural statistics and constraint violations (SI Appendix, Table S1) allowed us to identify the stereoisomer with the D stereochemistry at Ala12 (α-S), Asn16 (α-S), Arg34 (α-S), and Lys42 (α-S) as a representative structure given 1) the absence of thioether bridge constraint violations, 2) the great number of nOe connectivities used in the structure calculation, and 3) the lowest average target function value and a low rmsd. To improve the structure of the DDDD stereoisomer, an additional refinement step by returning to the Nuclear Overhauser Effect SpectroscopY (NOESY) spectra to eliminate the ambiguities found during the structure calculation by the CYANA software was added. The resulting structural statistics of the 20 conformers for the DDDD isomer of RumC1 are summarized in SI Appendix, Table S2. The backbones of the 20 lowest target function value conformers for the DDDD isomer of RumC1 superimpose quite well with an rmsd value of 0.81 Å for the backbone (Fig. 1B).” should instead appear as “The structural statistics and constraint violations (SI Appendix, Table S1) allowed us to identify the stereoisomer with the L stereochemistry at Ala12 (α-R), Asn16 (α-R), Arg34 (α-R), and Lys42 (α-R) as a representative structure given 1) the absence of thioether bridge constraint violations, 2) the great number of nOe connectivities used in the structure calculation, and 3) the lowest average target function value and a low rmsd. To improve the structure of the LLLL stereoisomer, an additional refinement step by returning to the NOESY spectra to eliminate the ambiguities found during the structure calculation by the CYANA software was added. The resulting structural statistics of the 20 conformers for the LLLL isomer of RumC1 are summarized in SI Appendix, Table S2. The backbones of the 20 lowest target function value conformers for the LLLL isomer of RumC1 superimpose quite well with an rmsd value of 0.81 Å for the backbone (Fig. 1B).” The online version has been corrected.
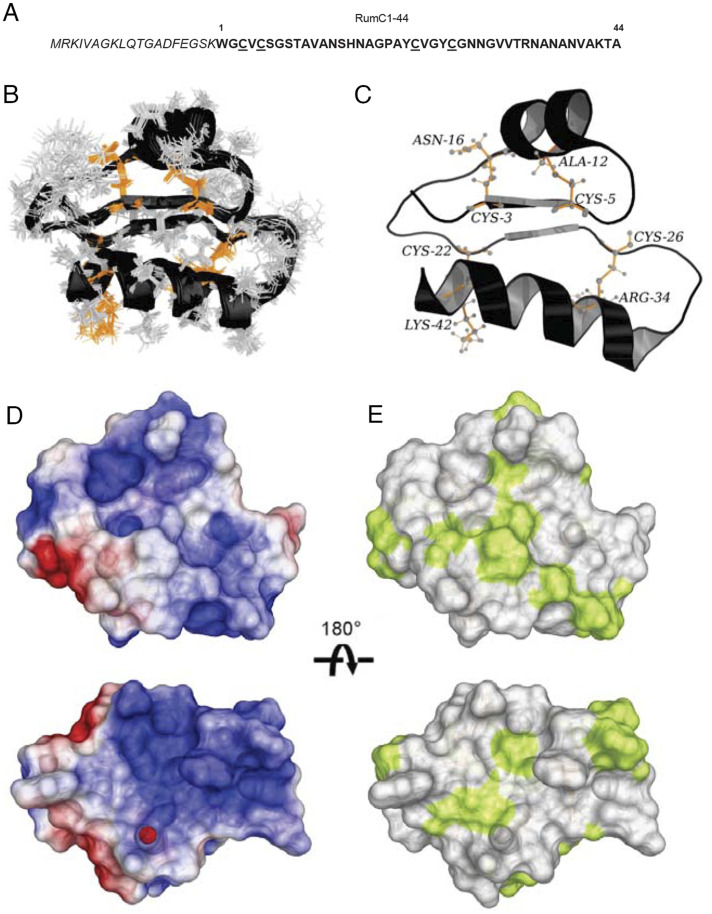
Fig. 1.
Sequence and three-dimensional structure of Ruminococcin C1. (A) Sequence of RumC1 containing leader peptide (italics) and core peptide (RumC1-44); cysteine residues are underlined. (B) Backbone overlay of the 20 lowest target function value conformers for the LLLL stereoisomer of RumC1. (C) Cartoon backbone representation of the three-dimensional solution structure of RumC1 with the L stereochemistry at Ala12 (α-R), Asn16 (α-R), Arg34 (α-R), and Lys42 (α-R). Cysteine sulfur to α-carbon thioether cross-links are colored in orange, and the position are indicated. (D) Electrostatic surface potential of RumC1, where blue indicates positive charge and red indicates negative charge. (E) Surface hydrophobicity of RumC1, where yellow represents hydrophobic residues and white represents hydrophilic residues.
The authors note that on page 19170, right column, first full paragraph, line 1, “The electrostatic surface potentials present an overall positive charge (Fig. 1D), and the surface hydrophobicity of the DDDD stereoisomer shows a majority of hydrophilic residues (Fig. 1E).” should instead appear as “The electrostatic surface potentials present an overall positive charge (Fig. 1D), and the surface hydrophobicity of the LLLL stereoisomer shows a majority of hydrophilic residues (Fig. 1E).” The online version has been corrected.
The authors note that on page 19170, right column, first full paragraph, line 6, “Four sulfur toα-carbon thioether bridges with a DDDD stereochemistry have been already reported for thurincin H (10).” should instead appear as “Four sulfur to α-carbon thioether bridges with a DDDD stereochemistry have been reported for thurincin H (10).” The online version has been corrected.
The authors note that on page 19174, left column, first full paragraph, line 8, “In contrast to a recent report by Berteau and coworkers, where the residues involved in the thioether bridges were identified to be L-configurated, after hydrolysis and derivatization (15), here we found that the stereoisomer that fits best with the NMR data featured D-configurations at Ala12, Asn16, Arg34, and Lys42.” should instead appear as “In agreement with a recent report by Berteau and coworkers, where the residues involved in the thioether bridges were identified to be L-configurated, after hydrolysis and derivatization (15), here, we found that the stereoisomer that fits best with the NMR data featured L-configurations at Ala12, Asn16, Arg34, and Lys42.”. The online version has been corrected.
The authors note that on page 19176, right column, third full paragraph, line 6, “Coordinates of the 20 conformations of DDDD stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.” should instead appear as “Coordinates of the 20 conformations of LLLL stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.” The online version has been corrected.
The authors note that the legend for Fig. 1 appeared incorrectly. The figure and its corrected legend appear below. The online version has been corrected.
The authors note that, in the SI Appendix, page 14, first full paragraph, line 10, “Coordinates of the twenty conformations of DDDD stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.” should instead appear as “Coordinates of the twenty conformations of LLLL stereoisomer of RumC1 have been deposited into the Protein Data Bank under accession code 6T33.” The SI Appendix has been corrected online.
The authors note that the legend for SI Appendix, Fig. S2 appeared incorrectly. The SI Appendix has been corrected online.
The authors note that SI Appendix, Tables S1 and S2 appeared incorrectly. The SI Appendix has been corrected online.