Figure 2.
Differences in the p110γ interface in p84 versus p101
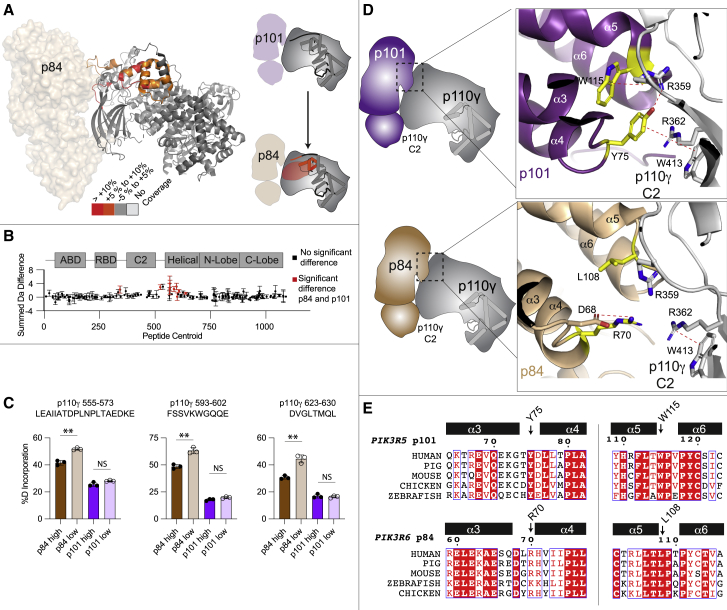
(A) HDX-MS differences in the p110γ subunit between the p110γ-p101 and p110γ-p84 complexes. Significant differences in deuterium exchange (defined as greater than 5%, 0.4 Da, and a two-tailed t test p < 0.01 at any time point) are mapped on to the structure of p110γ-p84 and cartoon of p110γ according to the legend.
(B) Sum of the number of deuteron differences between the p110γ-p101 and p110γ-p84 complexes over the entire deuterium exchange time course. Positive difference is indicative of enhanced exchange in p110γ-p84. Each point is representative of the center residue of an individual peptide. Peptides that met the significance criteria described in (C) are colored red. Error is shown as the sum of the standard deviation across all time points (n = 3 for each time point). All HDX-MS data are provided in the source data.
(C) Selected deuterium exchange at 30 s for peptides in p110γ for p110γ-p101 and p110γ-p84 complexes at either a high concentration (1,500 nM) or a low concentration (175 nM). Error is shown as standard deviation (n = 3) with two-tailed p values as indicated: ∗∗p < 0.01; not significant (ns) > 0.05. Full HDX-MS data for all peptides in this experiment are shown in the source data.
(D) Cartoon schematic of the p110γ interface for p101 (top) and p84 (bottom), with a zoom in on the residue’s located at the p110γ interface for both p84 and p101. Dotted lines indicate cation-pi or electrostatic interactions.
(E) Sequence alignment of both p101 and p84 residues in the α3 to α6 helices located at the p110γ interface. The residues annotated in panel are indicated on the alignment. A full alignment of p101 and p84 is shown in Figure S3.