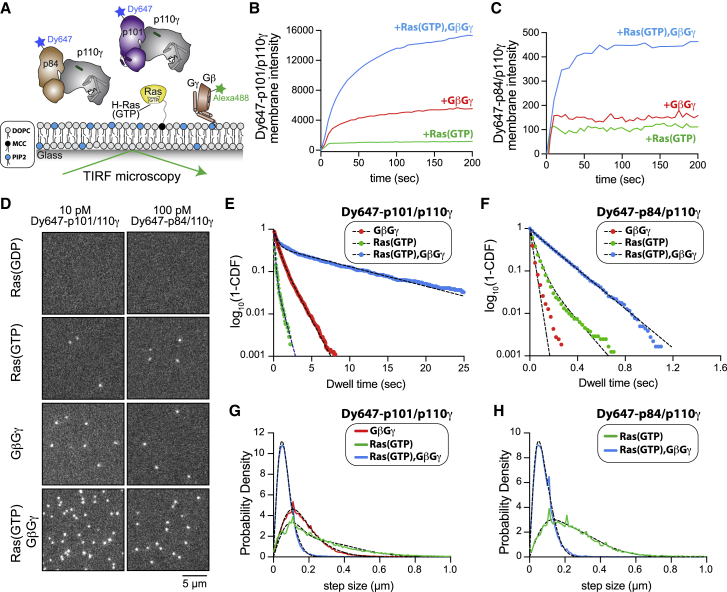
Figure 5.
p110γ-p84 and p110γ-p101 exhibit distinct membrane-binding dynamics in the presence of HRas and Gβγ
(A) Experimental setup for visualizing DY647-p101-p110γ and DY647-p84-p110γ interactions with a supported lipid bilayer containing membrane-anchored HRas(GDP or GTP) and farnesylated Gβγ. Experiments show a representative experiment, with all quantification generated from 2 to 4 technical replicates (see Table S3).
(B and C) Kinetics of PI3K complex membrane recruitment measured in the presence of 10 nM DY647-p101-p110γ or DY647-p84-p110γ using TIRF-M.
(D) Representative smTIRF-M images visualizing PI3K complex localization in the presence of 10 pM DY647-p101-p110γ or 100 pM DY647-p84-p110γ. Localization was measured on SLBs containing Ras(GDP), Ras(GTP), Gβγ, or Ras(GTP)/Gβγ.
(E and F) Single-molecule dwell time distributions for DY647-p101-p110γ or DY647-p84-p110γ measured in the presence of the indicated binding partners. Plots showing log10(1-cumulative distribution frequency [CDF]) versus dwell time (s). Data are fit to either a single or double exponential decay curve (black dashed lines). Single-molecule imaging of DY647-p84-p110γ yielded the following mean dwell times: 39 (+RasGTP), 74 (+Gβγ), and 188 ms (+RasGTP/Gβγ). Single-molecule imaging of DY647-p101-p110γ yielded the following mean dwell times: 0.146 (+RasGTP), 0.73 (+Gβγ), and 3.09 s (+RasGTP/Gβγ).
(G and H) Step size (or displacement) distributions of DY647-p101-p110γ or DY647-p84-p110γ. PI3K complex formation with Ras(GTP), Gβγ, or Ras(GTP)/Gβγ modulates the single-molecule displacement (i.e., step size, μm). Dashed black line represents the curve fit used to calculate the diffusion coefficient (see STAR Methods).
The membrane composition for membranes in (B)–(H) was 96% DOPC, 2% PI(4,5)P2, 2% MCC-PE. See Table S3 for time constants (τ1 and τ2), diffusion coefficients (μm2/s), and statistics (n = 2–4).