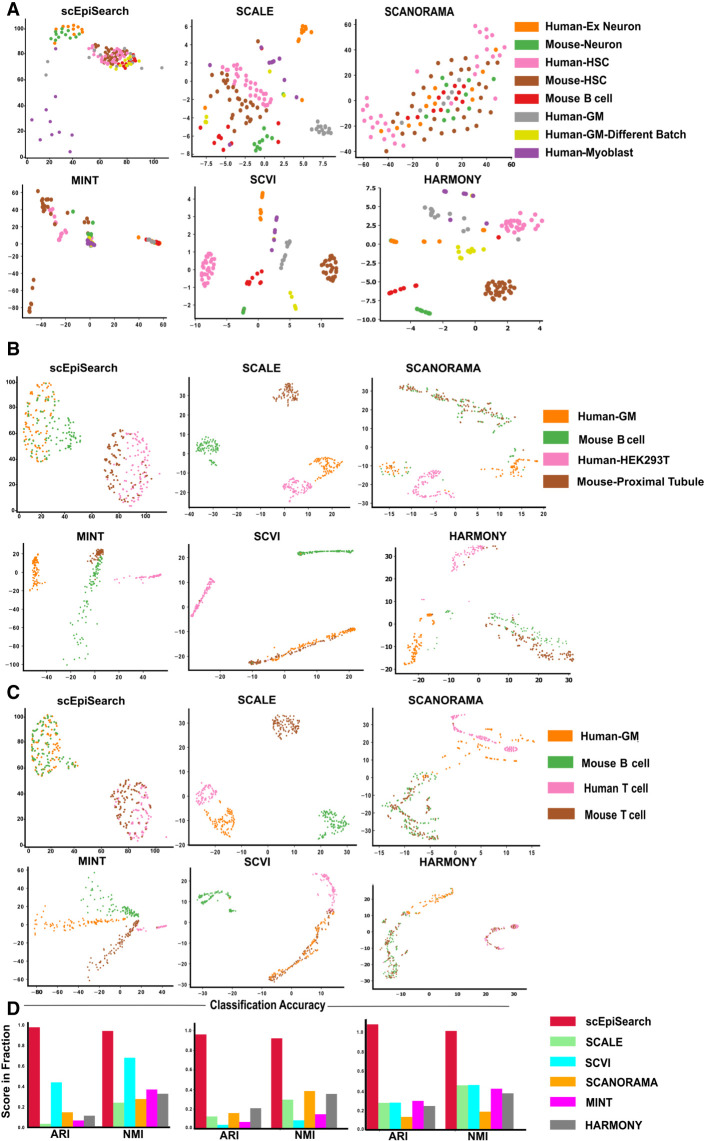
Figure 4.
Evaluation of embedding of query sets of single-cell open-chromatin profiles irrespective of batch effect, species, differences in peak list and their source (scientific group). (A) For this case study, queries consisted of separate read-count matrices for scATAC-seq profiles of human-neuron, mouse-neuron, human-HSC (hematopoietic stem cells), mouse-HSC, human-myoblast, human-GM12878 (GM) cells from two batches, and mouse B cells. Here the x-axis shows dimension-1 whereas the y-axis shows dimension-2 of low dimensional embedding. The peak lists of query read-count matrices were different from each other. Embedding plots from five other methods are also shown here. While SCANORAMA mixed the location of all cells, MINT could not group cells of the same type together like scEpiSearch. (B) The plot of embedding results shows the alignment of the same cells from different species and batches together. Queries are made for human-GM12878 (GM) cell, mouse B cell, human-HEK293T, and mouse-proximal tubule. The embedding plot from scEpiSearch derived from projections onto mouse expression profiles. (C) The plots show the 2D embedding of cells from different species and batches. Queries were made for human-GM12878 (GM), mouse B cell, human T cell, and mouse T cell. (D) The purity of density-based spatial clustering (using DBSCAN) with embedded coordinates is also shown here in terms of ARI and NMI scores. The silhouette coefficients calculated without DBSCAN-based clustering are shown in Supplemental Figure S12B.