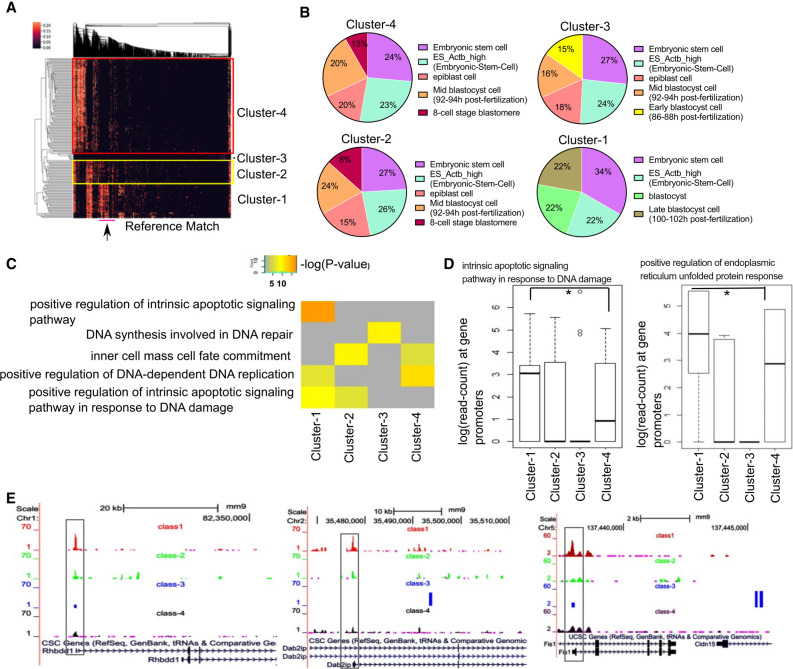
Figure 6.
Studying single-cell epigenome profile of stem cells using scEpiSearch. (A) Heatmap obtained from scEpiSearch showing biclustering of match scores of query scATAC-seq profiles of mouse embryonic cells (rows) with top matching reference single-cell epigenome profile (columns). The columns, highlighted with the pink bar below (and arrow), belong to reference cells from the late blastocyst, which show high similarity with the mESC of clusters-1. (B) The pie charts show cell types of the top five matching expression profiles to query scATAC-seq profiles of mESCs belonging to different clusters. It is based on a top-2000 enriched genes-based search. (C) The result of gene-set enrichment for selected biological functions using genes proximal to the top 10,000 peaks specific to mESC cells belongs to different clusters. The gene-set enrichment was performed using the GREAT Gene Ontology enrichment tool using default parameters. (D) The read counts at the promoter of genes belonging to the gene set for biological function terms: “intrinsic apoptotic signaling pathway in response to DNA damage” and “positive regulation of endoplasmic reticulum unfolded protein response.” Here a matrix consisting of a read count for four clusters is quantile normalized to avoid bias. The star (*) shows a significant P-value (<0.03) calculated using Wilcoxon rank-sum test. (E) The snapshot of the UCSC Genome Browser showing the difference in the activity level of the promoter of two genes (Rhbdd1, Dab2ip) associated with unfolded protein response and endoplasmic reticulum stress and one gene (Fis1) associated with apoptosis, cell cycle, and mitochondrion fission.