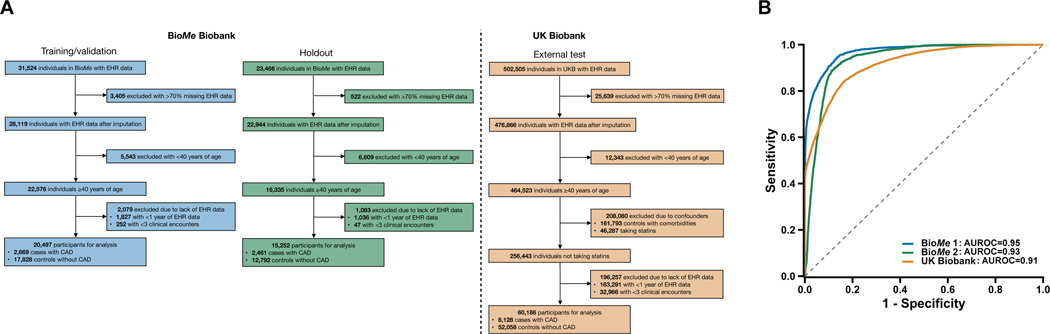
Fig. 1.
Performance of the machine learning model for the detection of coronary artery disease (CAD) in the validation, holdout, and external test sets.
The machine learning model was trained/validated in the BioMe Biobank (BioMe 1), assessed in a holdout set in BioMe (BioMe 2), and externally tested in the UK Biobank. a, Electronic health records (EHRs) of study participants contained both categorical data (i.e., diagnosis codes and medications) and continuous data (i.e., laboratory readings and vital measurements). Only EHR data prior to the earliest date of coronary artery disease (CAD) diagnosis, procedure (e.g., angioplasty), or medication (e.g., statins) prescription were used for CAD cases. In UK Biobank, date of statins prescription is unavailable and individuals with statins were excluded; controls with an Elixhauser comorbidity index of zero were retained. Participants with >70% missing data in the EHR were removed, and the EHR data of the remaining individuals underwent imputation with a random forest-based algorithm. We restricted to participants at least 40 years of age as the target population for which CAD is prevalent and the pooled cohort equations (PCE) is designed to guide statin initiation. Age was defined by the last considered clinical feature entry. Participants with at least one year of EHR data and three recorded clinical encounters were retained. b, The machine learning model discriminated CAD controls from cases with area under the receiver-operating-characteristic curves (AUROCs) of 0.95 (95% CI, 0.94–0.95), 0.93 (95% CI, 0.92–0.93), and 0.91 (95% CI, 0.91–0.91) for the validation, holdout, and external test datasets, respectively.