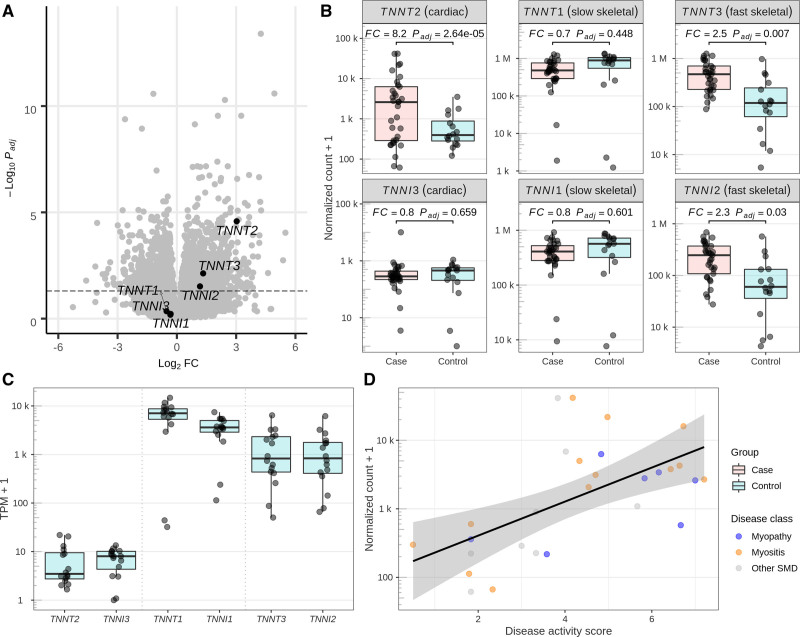
Figure 5.
mRNA analyses of muscle biospies. A, Differential gene expression (DGE) results from a case/control study comparing skeletal muscle biopsies from patients with (33 cases) and without (16 controls) skeletal muscle disorder (SMD). After correction for multiple testing with the Benjamini and Hochberg procedure, 847 of 17 124 protein-coding genes were upregulated and 966 were downregulated at a significance level of 𝛼 = 0.05. Three of 6 genes of the troponin gene family show a significant upregulation (TNNT2, top 96 differentially expressed gene [DEG]; TNNT3, top 881 DEG; TNNI2, top 1821 DEG). B, Detailed DGE results for 6 genes of the Troponin gene family. Fold changes (FCs) and significance levels are concordant among the slow (TNNT3 and TNNI2) and fast (TNNT1, TNNI1) skeletal muscle gene pairs but not for the cardiac gene pair (TNNT2 and TNNI3). C, Base-level expression of the troponin gene family in skeletal muscle. Cardiac genes TNNT2 and TNNI3 exhibit an expression of 6 and 6.3 transcripts per million (TPM), ranking among the top 39% and 38% expressed protein-coding genes in the DGE analysis. Fast and slow skeletal muscle genes TNNT1, TNNI1, TNNT3, and TNNI2 exhibit a mean expression of 6848, 3614, 1590, and 1418 TPM (all top 0.1%). D, Variation of TNNT2 expression in the case samples (n=28 after filtering for missingness in marker variables for disease activity) can be explained by biopsy-specific disease activity. Linear regression shows a significant positive correlation (R=0.59, P<0.001) between a disease activity score derived from 14 disease activity markers and normalized counts. The score remains significant (P=0.001) after adjustment for disease class (myopathy [n=7], myositis [n=13], other SMD [n=8]). Conversely, the case subset showed borderline significant differences between disease classes after adjustment for disease activity score in a likelihood ratio test (P=0.069). A detailed description of the score calculation is available in the Supplement Material.