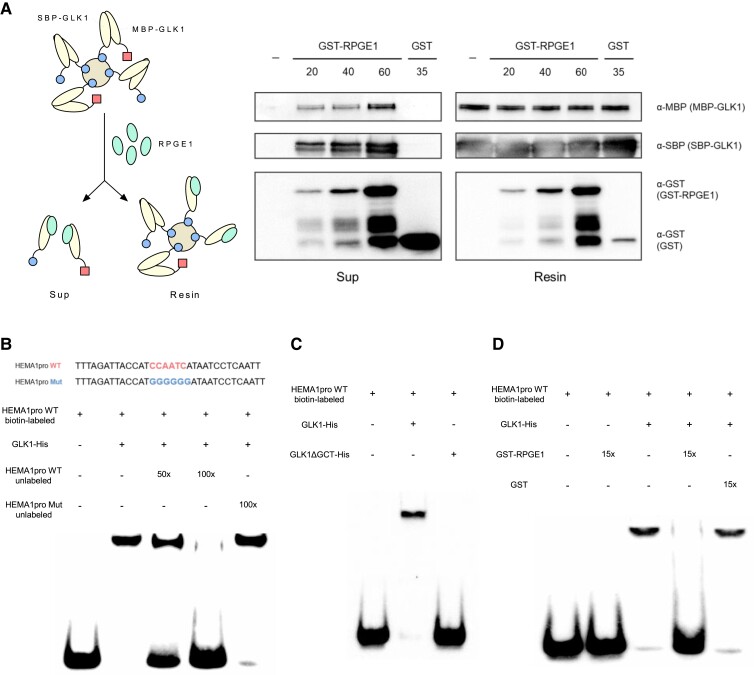
Figure 6.
RPGE1 inhibits the DNA binding of GLK1 by disrupting its dimerization. A, The disruption of Arabidopsis GLK1 dimer by Arabidopsis RPGE1 in vitro. A diagram in the left indicates the experimental scheme. RPGE1 (GST-RPGE1-His) was added to streptavidin resin-bound GLK1 dimers of SBP-GLK1/SBP-GLK1 or SBP-GLK1/MBP-GLK1. After elution, immunoblotting with the appropriate antibodies (α-GST, α-SBP, and α-MBP) was used to detect each protein in the supernatant (Sup) and resin. The addition of buffer only or GST instead of GST-RPGE1-His is indicated by the minus (−) sign and GST, respectively. B, The binding of Arabidopsis GLK1 to a HEMA1 promoter fragment, as determined by electrophoretic mobility shift assay (EMSA). Upper panel shows sequences of the HEMA1 WT and mutant probes. EMSA was performed with biotin-labeled HEMA1 probe. Competition for the binding was tested with 50 × or 100 × of unlabeled WT probe or 100 × of unlabeled mutant probe. C, The failure of the DNA binding of the dimerization domain-deleted Arabidopsis GLK1 (GLK1ΔGCT). The EMSA was performed as in (B) except in the presence of Arabidopsis GLK1ΔGCT. D, Inhibition of the DNA binding of Arabidopsis GLK1 by Arabidopsis RPGE1. EMSA was performed as described in (B) except in the presence of Arabidopsis GST-RPGE1 or GST. GST or GST-RPGE was added to a 15 × molar concentration of GLK1 protein.