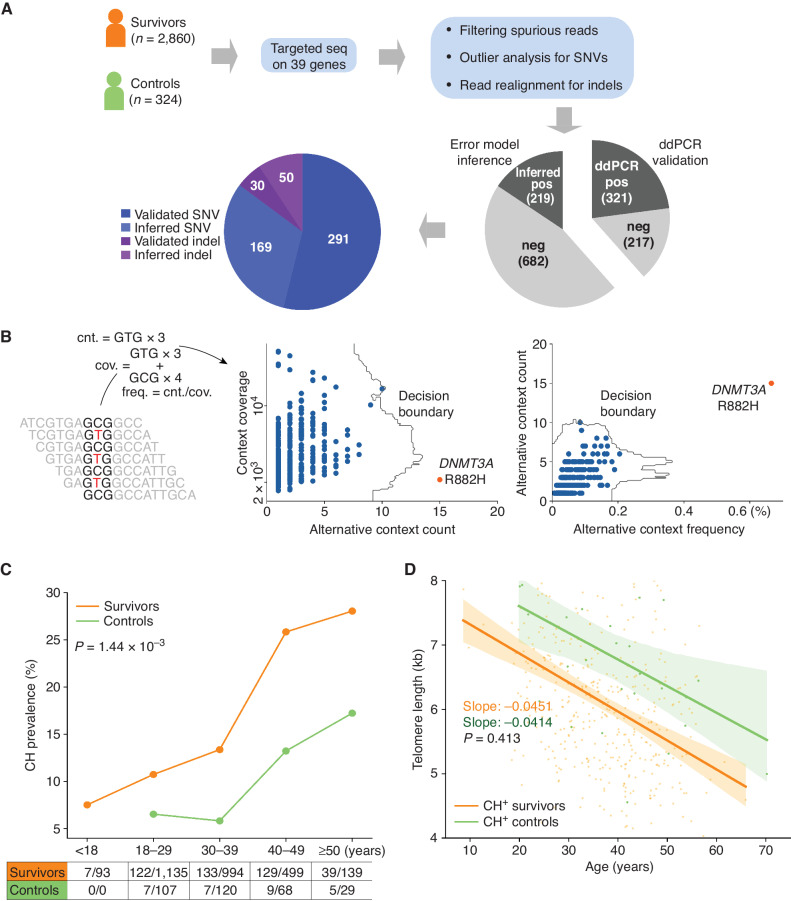
Figure 1.
Elevated CH in pediatric cancer survivors. A, Study design for CH analysis in survivors. Blood samples from survivors and controls were analyzed by capture sequencing on 39 CH-associated genes. Putative CH variant analysis involves computational error suppression of sequencing reads, outlier analysis of substitutions with the same genomic context, and indel realignment. Approximately 40% of the putative variants (538 total) were successfully assayed by ddPCR, whereas the validity of the remaining variants was inferred by comparing to a background error model. ddPCR, digital droplet PCR; SNV, single-nucleotide variant. B, Outlier analysis based on the sequence context of a CH variant. For each of the 96 genomic triplet context changes, the count and frequency for sequencing context matching the context of the alternative allele as well as the read count were within a sample. Outlier analysis was performed by IsolationForest (17) in both the spaces spanned by the count and coverage and count and frequency. Example shows the GCG>GTC context analysis in an acute lymphoblastic leukemia survivor, in which the DNMT3A R882H mutation was detected as an outlier. cnt., count; cov., coverage. C, Elevated CH prevalence in the survivors (orange) compared with the controls (green) across all five age categories. Significance of difference in the overall prevalence of the two groups was based on Fisher exact test. D, Leukocyte telomere lengths in the CH-positive survivors and controls. Telomere attrition rates were estimated as a regression slope, which was compared by the t test. Bands represent 95% CIs of the least-square regressions.