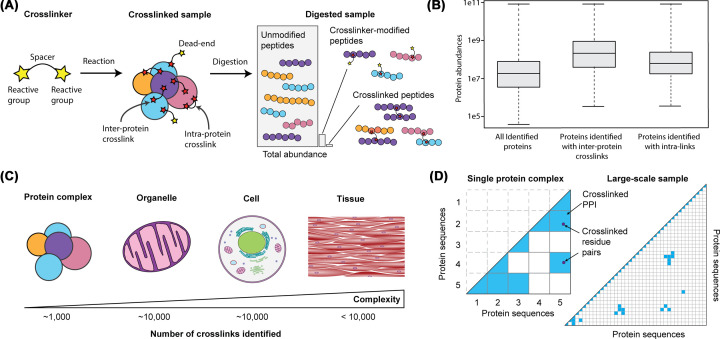
Figure 1. Challenges for identifying cross-linked peptides from complex samples.
(A) Cross-linkers are made up of two reactive groups and a spacer that dictates the maximum distance between the two cross-linked residues. After the cross-linked sample is digested the cross-linked peptides make up a small percentage of the total mass in the sample. (B) In large-scale samples containing 1000s of proteins cross-links have only been detected from the most abundant proteins. Intra-protein cross-links tend to be easier to identify than inter-protein cross-links. (C) All biological complexities have been studied with cross-linking MS but the coverage of the proteomes in all complex samples is poor. (D) The number of peptide combinations to be searched in purified complexes is tiny compared with all the possible combinations in samples containing thousands of proteins. This means that for these more complicated samples the fragmentation spectra must be more detailed to confidently identify the cross-linked peptides.