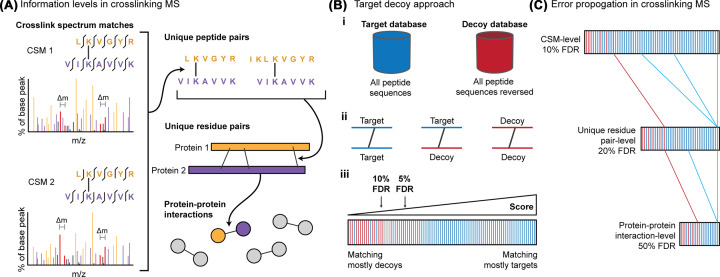
Figure 5. Error control for cross-linking MS.
(A) Cross-linking data contains multiple levels of information. Multiple CSMs can represent a single unique cross-linked peptide pair. Multiple cross-linked peptide pairs can represent a single unique residue pair and many unique residue pairs can represent one PPI. (B) i – The target-decoy approach is where each spectrum is searched against a ‘target’ database containing all the protein sequences, and against a ‘decoy’ database of false sequences of the same size and composition as the target database. ii – In cross-linking MS each CSM can be a combination of targets and decoys. iii – Matches to the decoy database are used as a proxy for noise and can be used to estimate the false discovery rate at any score threshold. (C) Error propagates through the information levels so the FDR should be estimated at the information level required for the study. False CSMs do not consolidate throughout the levels so if FDR is estimated at CSM-level, the error rate at PPI-level can be very high. The solution is to merge to the desired level (PPI-level, for examples) prior to calculating the FDR.