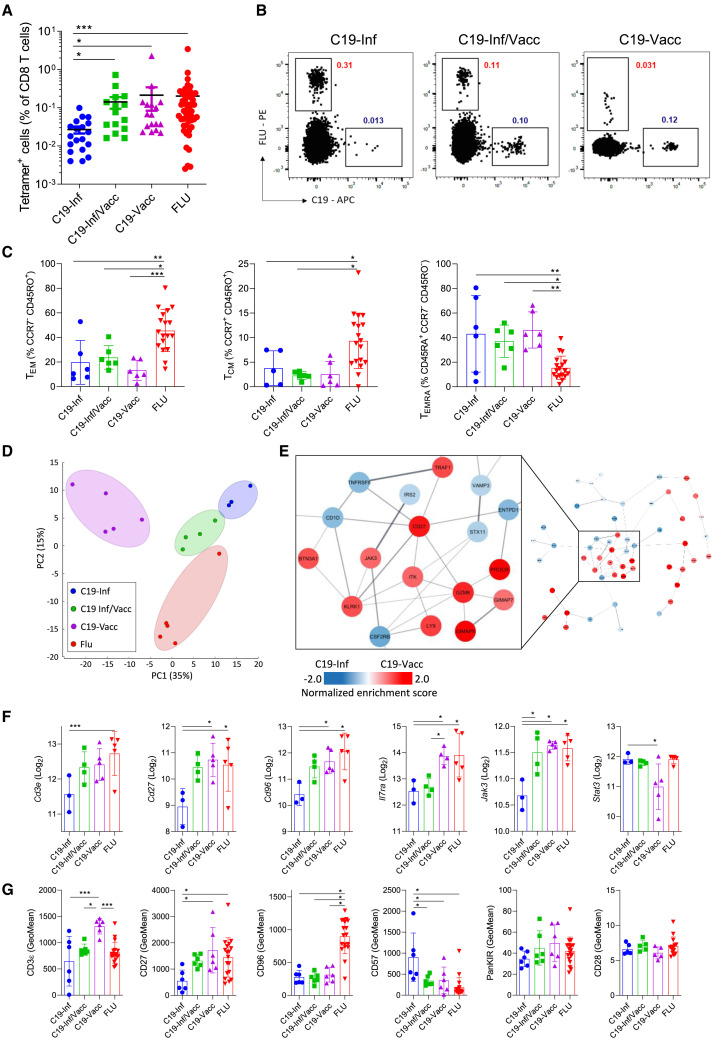
Figure 1.
The route of antigen exposure impacts the transcriptional and phenotypic profile of virus-specific memory cells
PBMCs from 3 groups of people were analyzed directly ex vivo (Inf, people with convalescent COVID-19; Inf/Vacc, people with convalescent COVID-19 who received 2 doses of vaccine; Vacc, nonconvalescent people who received 2 doses of COVID-19 vaccine).
(A) Frequency of antigen-specific (C19- or FLU-tetramer+) cells. Each dot represents one donor (n = 19 – C19-Inf, n = 15 C19-Inf/Vacc, n = 17 C19-Vacc, n = 51 FLU pooled from all 3 groups).
(B) Representative fluorescence-activated cell sorting (FACS) plots of cells stained with HLA A∗02 tetramers loaded with the YLQPRTFLL (C19) epitope of SARS-CoV-2 or GILGFVFTL (FLU) epitope of influenza. Gated is for live CD3+CD8+ cells.
(C) Quantification of TEM, TCM, and TEMRA cell subsets among C19- and FLU-specific CD8+ T cells (n = 18).
(D–F) C19- and FLU-specific CD8+ T cells were sorted and analyzed by RNA sequencing.
(D) Principal-component analysis of virus-specific cells based on all differentially expressed genes (n = 17).
(E) The 200 most differentially expressed genes between C19-tetramer+ cells from the C19-Inf and C19-Vacc groups were subjected to protein network clustering. Shown is the largest node network for each comparison. Inset shows the largest subcluster.
(F) Differential expression of individual genes (n = 17).
(G) Geometric mean fluorescence intensity (GeoMean) of selected markers on C19- and FLU-specific CD8+ T cells by flow cytometry (n = 18). Indicated are means ± SEM.
Statistical significances at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Kruskal-Wallis rank-sum test with Dunn’s post hoc test for multiple comparisons (A) or one-way ANOVA followed with Bonferroni post-testing (C, F, and G).
See also Figure S1.