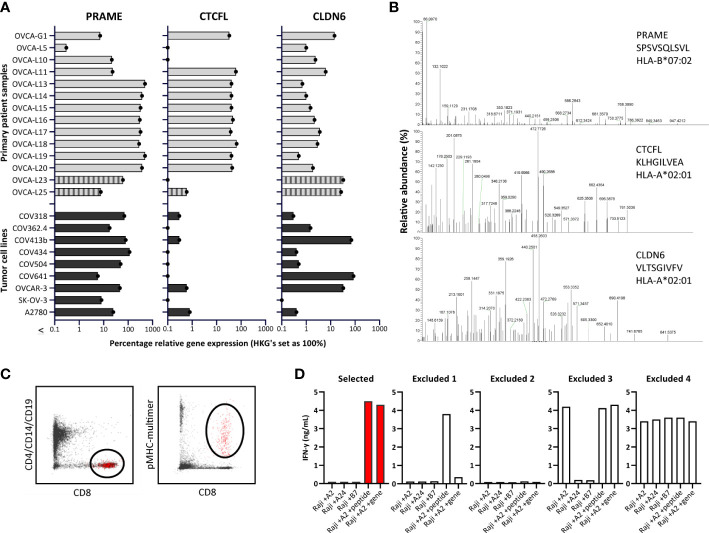
Figure 2.
Identification of PRAME, CTCFL and CLDN6 peptides and T-cell clones. (A) PRAME, CTCFL (TvX) and CLDN6 mRNA gene expression in 14 OVCA patient samples (12 solid tumor tissues and 2 malignant ascites samples (OVCA-L23 and OVCA-L25)), and 9 OVCA cell lines. Expression was measured by qPCR and is shown as percentage relative to the three HKGs GUSB, VPS29 and PSMB4, which was set at 100%. (B) Example of three OVCA-derived peptides identified in our HLA ligandome analyses. Shown are the mass spectra of the eluted peptides, including the gene, peptide sequence and HLA restriction. All eluted peptides were validated by comparing tandem mass spectra of eluted peptides and synthetic peptides, as shown in Supplementary Figure 3 . (C) Representative flow cytometry plots of the pMHC-multimer enriched cell population in 1 of the 25 healthy donors. Shown is the gating strategy of the single-cell sorted population (depicted in red), gated on CD8 (Alx700) +, pMHC-multimer (PE) + and CD4/CD14/CD19 (FITC) -. (D) Examples of recognition patterns based on IFN-γ production (ng/mL) of selected and excluded T-cell clones during the first T-cell screenings. T-cell clones were cocultured with Raji cells transduced with various HLA alleles, combined with loading of OVCA peptides (1 μM) or transduction of OVCA genes (E:T=1:6). Excluded 1 – 4 represent T-cell clones lacking potency and/or specificity. (HKGs, housekeeping genes; OVCA, primary ovarian cancer sample).