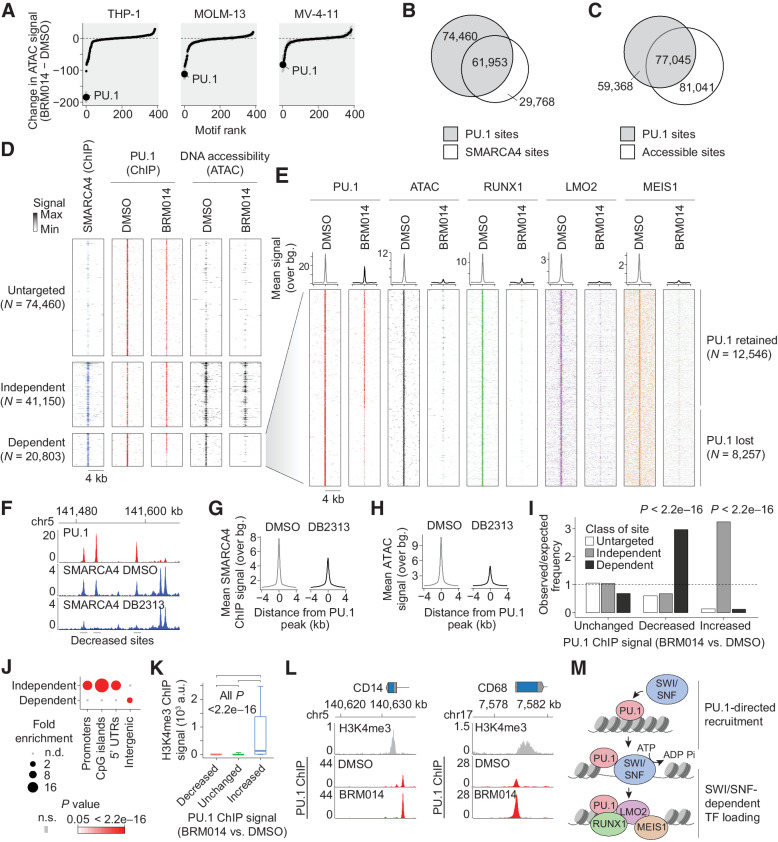
Figure 1.
SWI/SNF-mediated chromatin remodeling is required for PU.1-directed binding of AML CRC members. A, Ranking of differential TF motif accessibility in AML cell lines treated with 1 μmol/L BRM014 versus DMSO control. B, Overlap of PU.1 and SMARCA4 binding in THP-1 cells. C, Overlap of PU.1 binding and DNA accessibility in THP-1 cells. D, SMARCA4 occupancy and DNA accessibility at PU.1 sites in THP-1 cells treated with BRM014 or DMSO control. E, Binding of AML CRC factors RUNX1, LMO2, and MEIS1 at SWI/SNF-dependent PU.1 sites. F, Representative browser track of SMARCA4 binding at PU.1 sites upon DB2313 treatment. G and H, SMARCA4 binding (G) and chromatin accessibility (H) at PU.1 sites in THP-1 cells treated with DB2313. I, Enrichment of SWI/SNF-independent, -dependent, and untargeted sites at each class of PU.1 site based on altered PU.1 binding upon treatment with BRM014. Increased, decreased, and unchanged peaks in I and K correspond to PU.1 ChIP-seq changes regardless of any other feature. J, Enrichment of genomic features among SWI/SNF-dependent and -independent sites. K, H3K4me3 signal at sites with decreased, increased, or unchanged PU.1 occupancy upon BRM014 treatment. Box plot error bars indicate 10 percentile and 90 percentile range. L, PU.1 occupancy at the promoters of differentiation-related genes in BRM014- and DMSO-treated cells. M, Model of PU.1-directed recruitment of SWI/SNF to enhancer sites.