Figure 5.
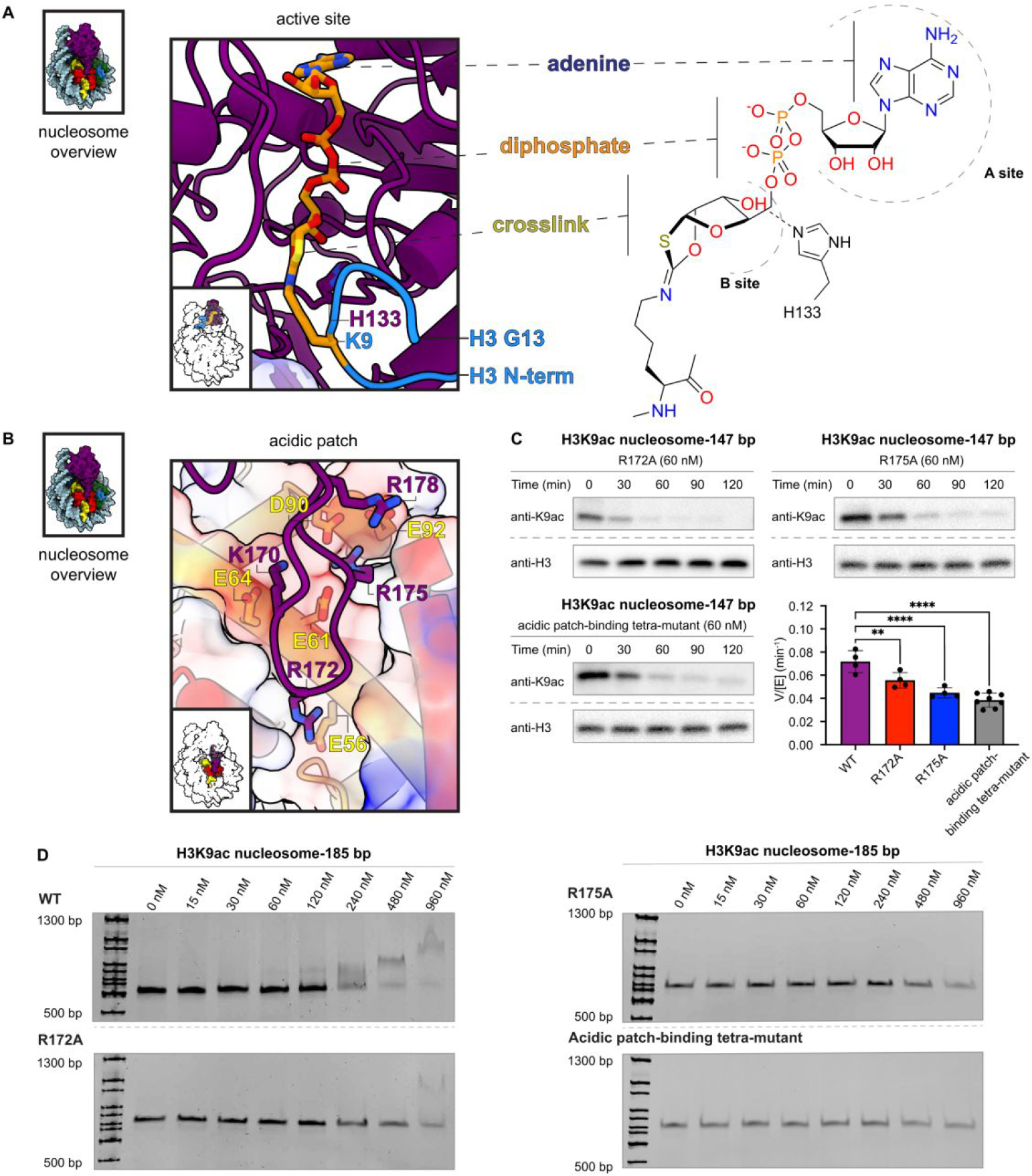
Cofactor crosslink and interactions between Sirt6 and the H2A/H2B acidic patch. (A) Sirt6 active site with modelled K9 Nε−1,3-oxathiolan-2-ylidene amine linkage to ADP-ribose. Histone H3 T3 through G13 are modelled (blue) based on an existing crystal structure (PDBID: 5Y2F).13 (B) Residues involved in interaction between Sirt6 (K170, R172, R175, R178) and H2A (E56, E61, E64, D90, E92). (C) Effect of point mutations to the acidic patch-binding loop on Sirt6 deacetylation of H3K9ac nucleosomes. R172A (top left, n=4), R175A (top right, n=4), and K170A/R172A/R175A/R178A (bottom left, n=8) each significantly decreased (** p=0.0089; **** p<0.0001) using an ordinary one-way ANOVA with multiple comparisons with the WT Sirt6 deacetylation rate (bottom right, n=4). (D) Electrophoretic mobility shift assay of acidic patch-binding point mutants. WT Sirt6 (top left) induces a change in nucleosome migration at the final two concentrations (480 nM and 960 nM). R172A (lower left), R175A (upper right) and K170A/R172A/R175A/R178A (bottom right) show only a slight change in nucleosome intensity at the final concentration (960 nM).
