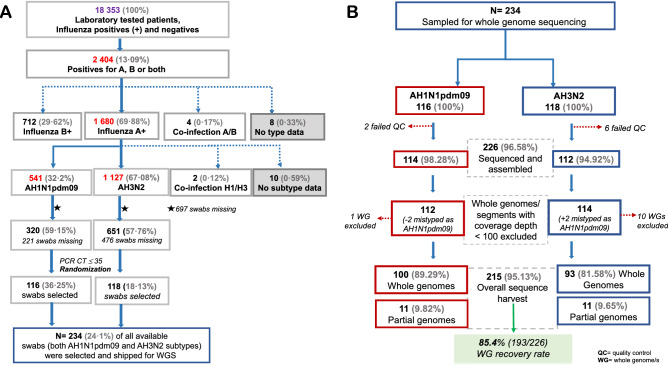
Figure 1.
Workflow of swab selection and whole genome recovery. (A) Shows how swabs were selected for influenza whole-genome sequencing (WGS). Patients diagnosed with either influenza subtypes A(H1N1)pdm09 or A(H3N2) and whose swab had a PCR CT ≤ 35 had their laboratory codes randomised based on the subtype and year of collection using the R software v3.6.3 (https://www.r-project.org). All available swabs were retrieved for years with less than fifteen swabs. The 697 swabs missing include some shipped to the Centers for Disease Control and Prevention (CDC) for routine surveillance and some lost due to an accidental failure of a freezer. The numbers are based on the UVRI-NIC laboratory dataset only, as of 9th May 2018. (B) Shows how viral samples were excluded before and after sequencing and the rate of whole genome recovery. Eight viruses [2 A(H1N1)pdm09 and 6 A(H3N2)] failed quality control (QC) before sequencing.