Figure 1.
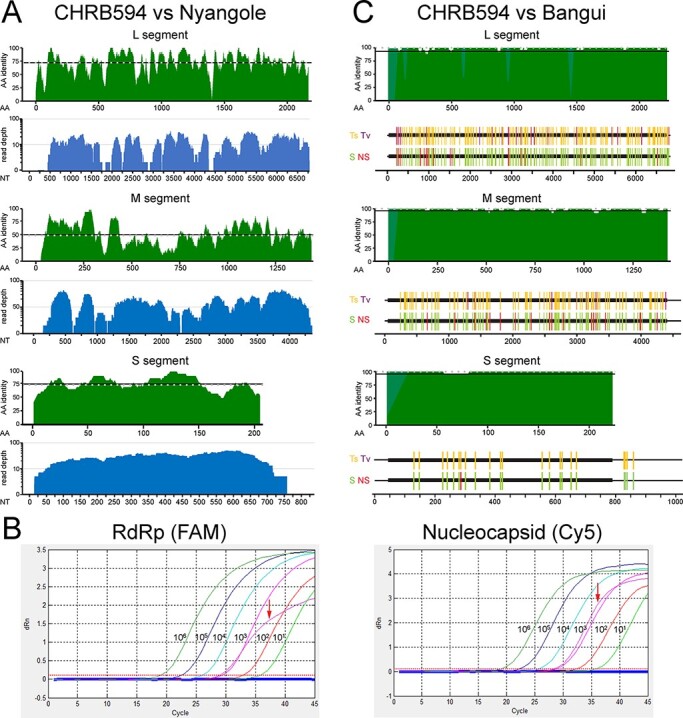
The independent discovery of a second Bangui virus strain. (A) Top (green): protein alignments performed between each genome segment of CHRB594 and Nyangole virus were scanned in twenty-five amino acid windows of the open reading frames. Dashed lines indicate the median per cent identity. Bottom (blue): NGS genome coverage plots for CHRB594. (B) RT-qPCR linearity for the L (left) and S (right) segments. The CHRB594 index case (bold curve) was detected in both channels at approximately Ct = 28, corresponding to a viral load of 3.2 log copies per liter. (C) Top (green): protein alignments between each genome segment of CHRB594 and Bangui virus (accession numbers: MK896632.1, MK896631.1, and MK896630.1). Gaps in alignment from the missing sequence in CHRB594 were filled with light green. Bottom: nucleotide alignments of full-length sequences highlighting transitions (gold) and transversions (purple), silent mutations (green), and non-silent mutations (red) in CHRB594 relative to Bangui virus.
