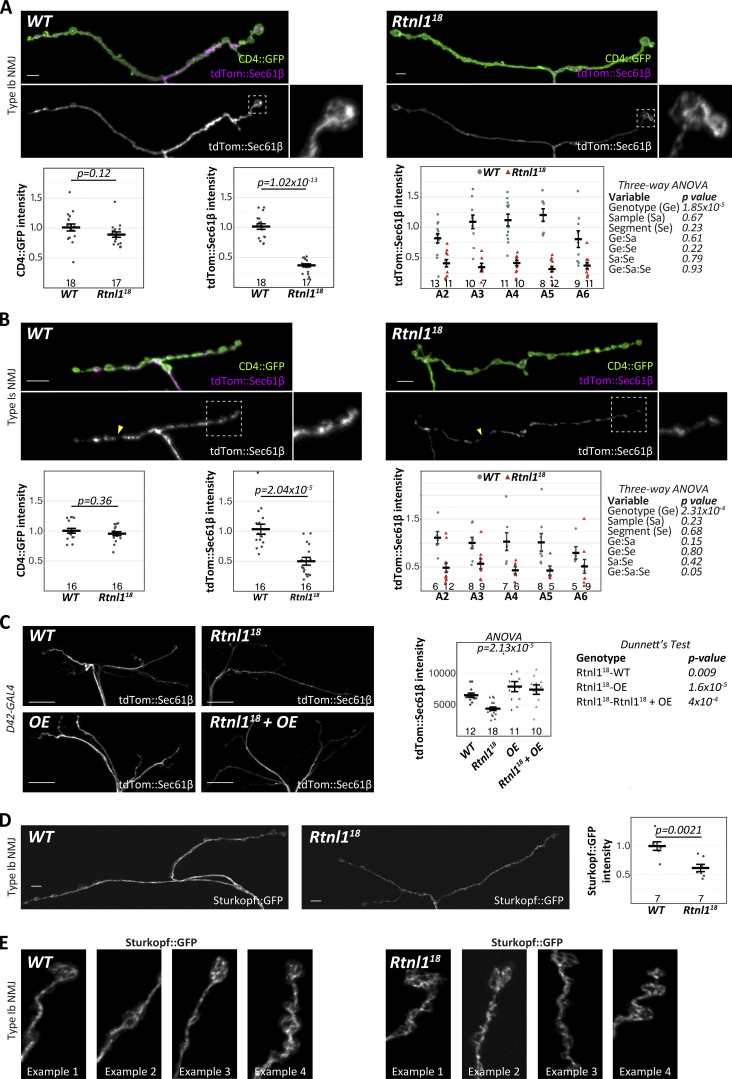
Figure 2.
Rtnl1 loss depletes presynaptic ER membrane markers. (A and B) Representative confocal projections and quantifications of ER marker tdTom::Sec61β in (A) Type Ib terminals on muscle 1 and (B) Type Is terminals on muscles 1–9 of WT and Rtnl118 larvae. Insets show magnified views of the areas inside broken lines. Arrowheads in B indicate gaps in tdTom::Sec61β signal. (C) Rtnl1::HA overexpression (OE) rescues the depletion of tdTom::Sec61β in Rtnl118 mutant NMJs at muscle 1/9. Since Type Ib and Is GAL4 constructs, UAS-Rtnl1::HA, and UAS-tdTom::Sec61β, are all inserted at attP2, we used D42-GAL4 which could be recombined with the UAS insertions. (D and E) Sturkopf::GFP in Type Ib muscle 1 NMJs, with intensity analyzed as above, and high magnification examples. All plots show individual larval datapoints and mean ± SEM; y-axes indicate arbitrary units (au) after normalization to WT; sample size is within the plots for each genotype. Where tdTom::Sec61β signal is compared between segments (A and B), sample size is the number of hemisegments (NMJs); for other plots, sample size is the number of larvae. For each larva, several NMJs across segments A2-A6 were randomly analyzed, and the mean value used as a larval datapoint. For each larva in C, we analyzed (but did not distinguish) Type Ib and Is branches on muscles 1 and 9 in a single segment chosen randomly between A4-A6. Student’s t tests were used for pairwise comparisons; ANOVA was used as shown for comparisons of more than two categories, or of multiple factors, with Dunnett’s post-hoc testing where appropriate. Scale bars for A, B, and D are 5 μm; for C, 20 μm. Insets in A and B are 8 × 8 μm. Panel width in E is 10 μm. Genotypes: A and B, GAL4, UAS-CD4::tdGFP/UAS-tdTom::Sec61β; C, D42-GAL4, UAS-tdTom::Sec61β and D42-GAL4, UAS-tdTom::Sec61β/UAS-Rtnl1::HA in either Rtnl1+ (WT) or Rtnl118 background (OE and Rtnl118 + OE respectively); D–E, Ib-GAL4, UAS-Sturkopf::GFP/+; all in either a WT or Rtnl118 background.