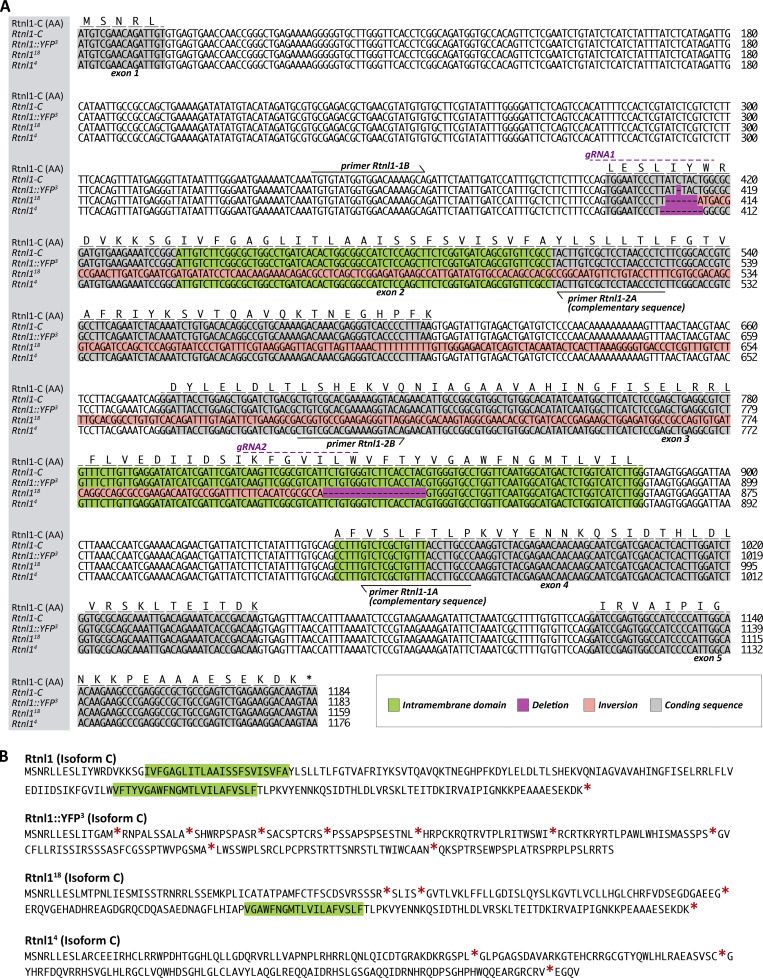
Figure S1.
CRISPR-derived lesions in Rtnl1 and their effects on the Rtnl1 coding region. (A) Manual alignment of the Rtnl1 transcript C for WT (Rtnl1-C), and Rtnl1::YFP3, Rtnl118 and Rtnl14 mutant CRISPR alleles, showing the amino acids (AA) encoded by the WT (Rtnl1-C) allele, the location of the gRNAs used to generate the mutant alleles, and the primers used to genotype them (see Materials and methods for details). Since the sequences encoding intramembrane domains are shared by all Rtnl1 isoforms, the shortest one, isoform C, was chosen for convenience. Sequences read from 5′ to 3′. The Rtnl1::YFP3 allele was generated from Rtnl1::YFPCPTI00291, has a 1-bp deletion/frameshift at the position of gRNA1 upstream of the first intramembrane domain, and lacks detectable YFP expression (Fig. S2); we did not use it further in this work, but it shows that frameshifts around this position can lead to loss of protein expression. The effects of Rtnl118 on the protein-coding sequence are described in the main paper. Rtnl14 has an 8-bp deletion/frameshift at the position of gRNA1, upstream of the first intramembrane domain. (B) Predicted Rtnl1-C protein sequences for the Rtnl1 alleles shown in A. Intramembrane domains are decorated in green and stop codons shown with a red asterisk.