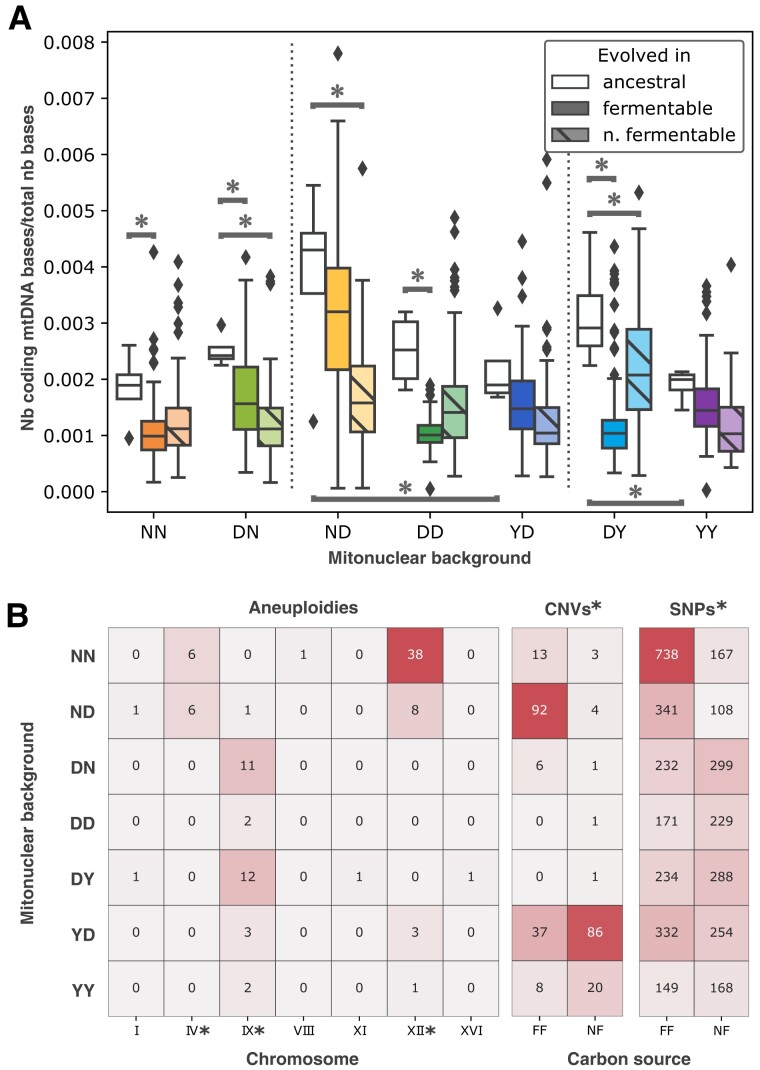
Fig. 3.
Whole-genome resequencing reveals mitonuclear background- and carbon source-specific genomic changes in experimental evolution isolates. (A) Fraction of sequenced bases mapped to coding regions of the mitochondrial genome was used as an indicator of genome-scale alterations to mtDNA, revealing impacts of mitonuclear background and evolution. Vertical dotted lines segregate strains based on mitochondrial background. Two-way ANOVAs comparing mtDNA fraction among strains bearing the same mitochondrial background reveal a significant effect for the nuclear background and evolution regimen (P < 0.05). Connectors marked by asterisks indicate significant differences in mtDNA fraction between ancestral strains bearing the same mitochondrial genome or between ancestral strains and their offsprings (Tukey HSD P < 0.05). (B) Changes in chromosome copy numbers (aneuploidies) and smaller, gene-scale copy number changes (CNVs) were detected using sequencing depth of coverage. Many single-nucleotide changes (SNVs) were also called. Asterisks indicate uneven distribution between mitonuclear backgrounds and carbon sources (χ2 test P < 0.05).