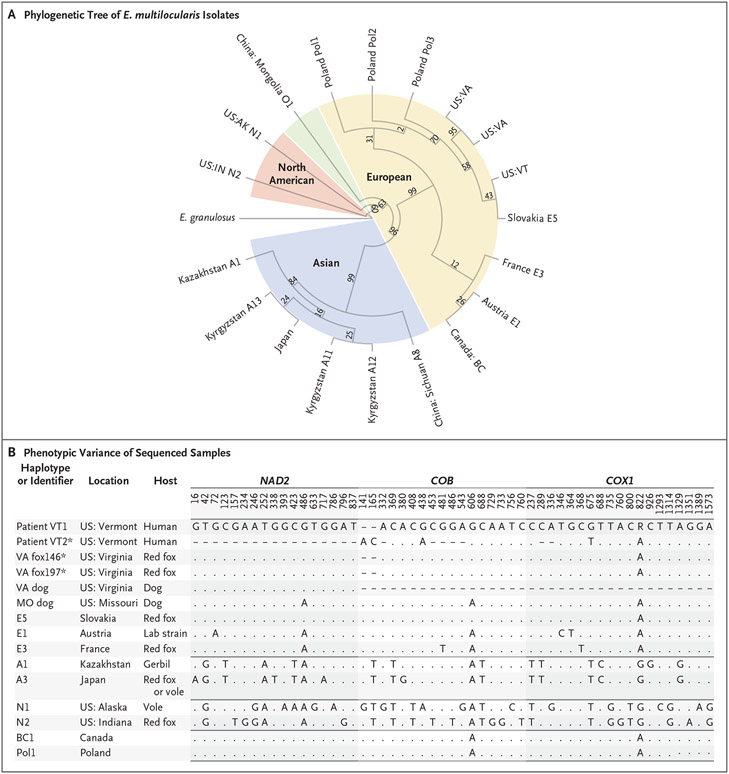
Figure 1. Phylogenetic Tree of Echinococcus multilocularis Isolates and Single-Nucleotide Polymorphisms (SNPs) That Distinguish among International Haplotypes.
Panel A shows the unscaled phylogenetic tree of the sequencing of E. multilocularis samples for NAD2, COB, and COX1, as reported previously and in the current study. The tree was constructed with the use of the maximum likelihood method. The isolate from a liver-biopsy sample obtained from Patient VT1 is indicated as US:VT, and two isolates from stool samples obtained from red foxes in Virginia are indicated as US:VA. Values were obtained by means of bootstrapping techniques performed with 1000 iterations. E. granulosus is shown on the left of the tree as an outgrouping. Panel B shows SNPs in NAD2, COB, and COX1 that most discriminate among Asian, European, and North American E. multilocularis haplotypes. Listed are most of the isolates that are shown in Panel A for comparison alongside all reported isolates from the United States that are consistent with a European strain. Dashes indicate that no data were available, and periods indicate that the nucleotide was identical to that in the lead sequence. Asterisks indicate that the sequences are reported in this study. Only NAD2 and COB sequences are reported in this study for Patient VT1.