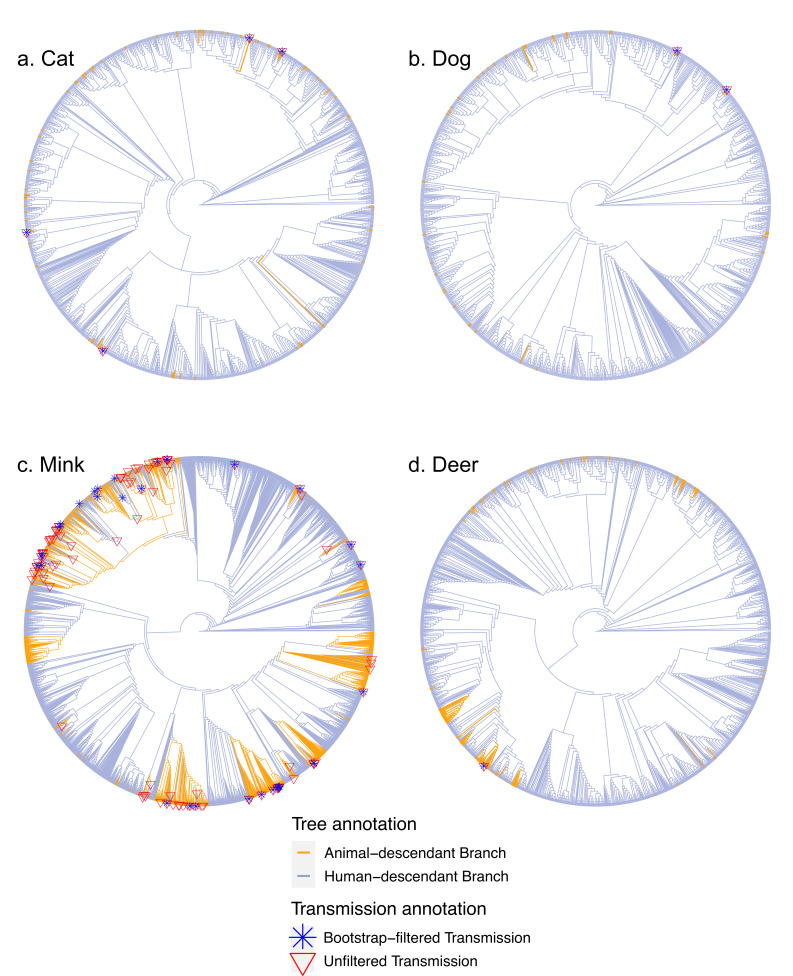
Figure 2. Transmission events inferred from non-human animals to humans.
Panels a-d display a representative tree for every species with animal-to-human transmissions marked on the tree. More detailed versions of these trees are in . Trees are rooted with the Wuhan reference genome (from one of the first sampled human COVID-19 patients).