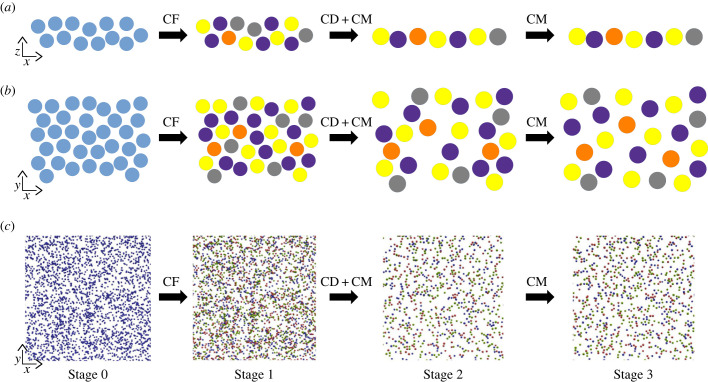
Figure 3.
Time course and spatial structure of a simulation using CF, CD and CM mechanisms. (a,b) schematic course of a simulation in x,z and x,y orientation, respectively. Undifferentiated cells are represented in blue and differentiated cells (RGC types) in yellow, indigo, grey and orange. For clarity purpose, only 4 out of the 43 implemented types are represented. (c) Time lapse of mosaic formation using BioDynaMo. Stage 0: First step of the simulation. Stage 1: Simulation after the end of CF mechanism, at day 1 (step 180). Average RI = 2.41 ± 0.1. Stage 2: Simulation after the end of CD mechanism, at day 6 (step 600). Average RI = 3.42 ± 0.31. Stage 3: Simulation at the end of the CM mechanism, at day 14 (step 2240). Average RI = 3.99 ± 0.36. Undifferentiated cells and represented in blue. On cells are represented in green, Off cells in red and On-Off cells in purple. The On, Off and On-Off groups are not RGC types but groups of types and thus do not form mosaics at the group level while each RGC type composing the group forms an individual mosaic. If CF mechanism is not simulated, simulations start at stage 1 with a defined cell type attributed at cells creation, accordingly to their type's theoretical density.