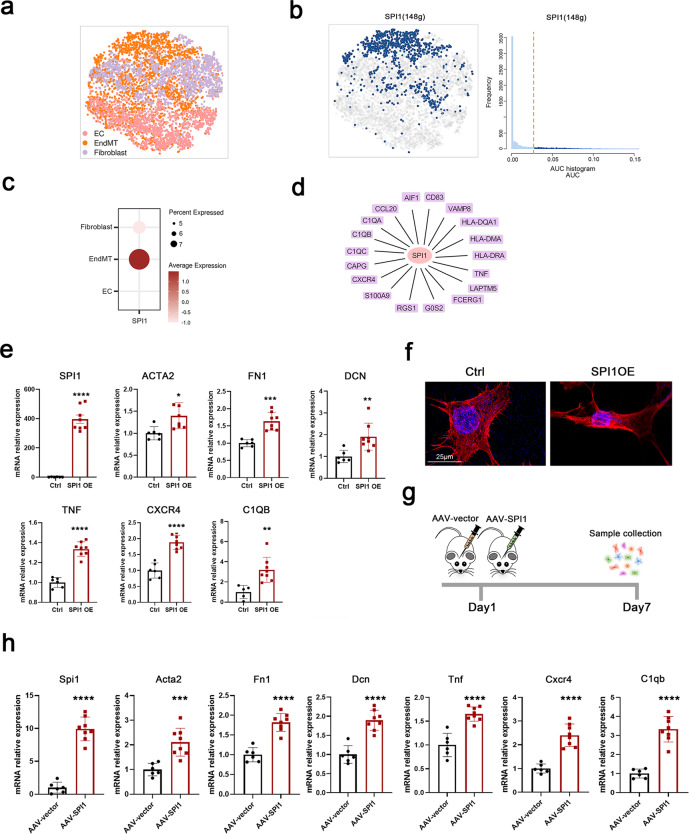
Fig. 5. SPI1 plays a key role in EndMT.
a SCENIC-based t-distributed stochastic neighbor embedding (tSNE) plot for the distributions of ECs, EndMT cells and fibroblasts. b SCENIC analysis predicted the transcription factor SPI1 as a specific hub (distinguished from ECs and fibroblasts) governing the EndMT cell state (left panel). SPI1 regulon activities were quantified using AUCell (right panel). c Dot plot for SPI1 expression in ECs, EndMT cells and fibroblasts. d Representative target genes of SPI1 overlapping with dynamic immune genes. e Expression levels of SPI1, EndMT markers (ACTA2, FN1 and DCN) and immune markers (TNF, CXCR4 and C1QB) regulated by SPI1 analyzed via real-time quantitative PCR (RT‒qPCR) assay in HUVECs after SPI1 and control plasmid transfection. f Morphological changes and cytoskeletal reorganization of HUVECs are shown by rhodamine-phalloidin staining (red) in the SPI1 overexpression and control groups. DAPI-stained nuclei are shown in blue. Scale bars, 25 µm. g SPI1 overexpression in the ECs of mice was induced by the injection of AAV-SPI1 with the TIE promoter stereotactically into the basal ganglia. Primary cerebrovascular ECs were collected 7 days after AAV-SPI1 and AAV-vector injection. h Expression levels of SPI1, EndMT markers (Acta2, Fn1 and Dcn) and immune markers (Tnf, Cxcr4 and C1qb) regulated by SPI1 analyzed via RT‒qPCR assay in primary ECs of cerebrovascular after AAV-SPI1 and AAV-vector injection. Primary ECs were isolated following the standard procedure of the MicroBeads kit and then used for RNA extraction and RT‒qPCR assay without culturing. Data are represented as the mean ± SD (AAV-vector group, n = 6; AAV-SPI1 group, n = 8). Statistics were performed using Student’s t test, and significance was determined as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.