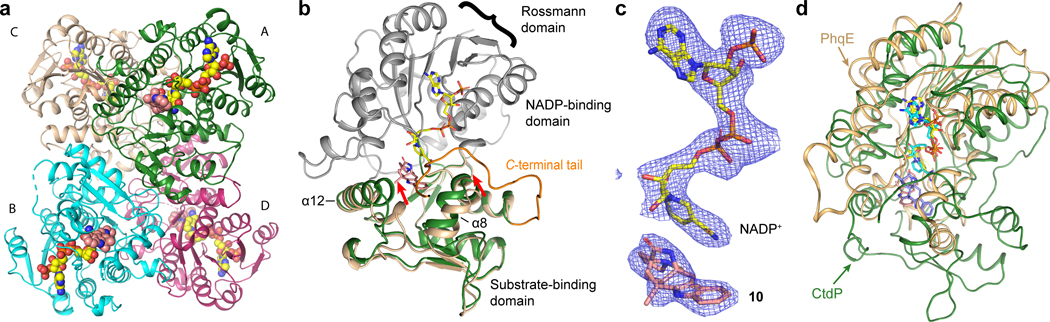
Fig. 2 |. Crystal structure analysis of CtdP complex.
a, Overall structure of the CtdP asymmetric unit is shown in a cartoon model and colored by each chain. Ligands are represented by spheres with NADP+ in yellow and penicimutamide E (10) in salmon. b, Alignment of CtdP chains A and C are shown as a cartoon, with NADP-binding domain in grey and substrate-binding domain forest green (chain A) or pale yellow (chain C). NADP+ (yellow) and 10 (salmon) from chain A shown as sticks. The C-terminal tail (residues 329–346) is highlighted with bright orange. c, The simulated annealing omits maps (blue mesh, Fo–Fc, contoured at 3.0 σ) of the NADP+ and 10. d, Superimposed CtdP-NADP+−10 (forest green) and PhqE-NADP+-(+)-11 (orange, PDB 6NKK) complex structures aligned by the NADP-binding domain.