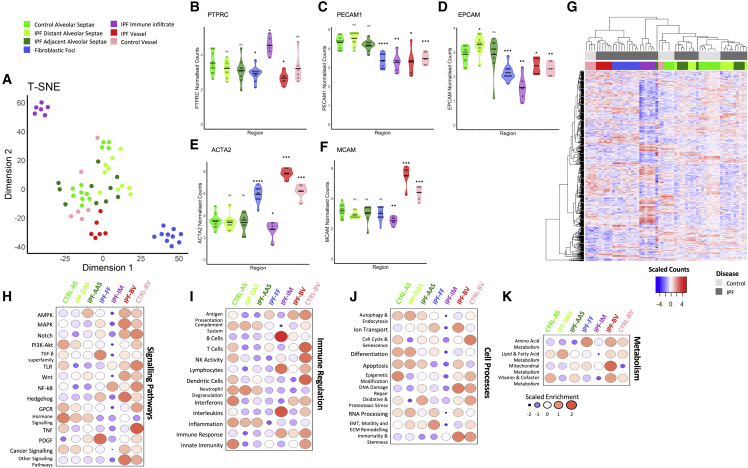
Figure 2.
Distinct gene expression signatures in spatially resolved regions of lung tissue
(A) t-Stochastic nearest neighbor embedding (t-SNE) dimensional reduction plot of each region of interest (ROI).
(B–F) Violin plots of gene expression values of cell population marker genes. Statistical comparisons are relative to control alveolar septae. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by Wilcoxon test with Benjamini-Hochberg multiple test correction.
(G) Heatmap showing differentially expressed genes (as measured by a Kruskal-Wallis test; p < 0.05) across dataset, showing clustering of different ROI groups.
(H–K) Bubble plots for control and IPF ROIs showing scaled enrichment scores for (H) signaling pathways gene sets, (I) immune regulation gene sets, (J) cell processes gene sets, and (K) metabolism gene sets.