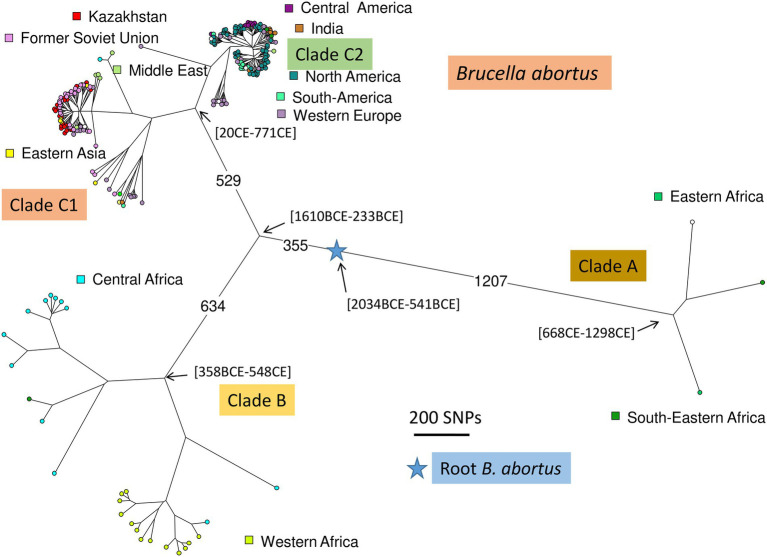
Figure 5.
Global B. abortus phylogeny. Maximum parsimony tree based on core genome SNPs. A total of 236 representative strains was used and 16,987 SNPs were called by mapping on genome accession GCA_000740155 (B. abortus strain Tulya). The size of the resulting tree is 17,178 SNPs (homoplasia 1.12%). Nodes are colored according to geographic origin as indicated. The blue star indicates the root of the phylogeny (branching point toward the B. melitensis type strain 16 M). The largest branch lengths and the scale are shown. Representative estimated divergence dates are indicated (CE, common era; BCE, before common era).