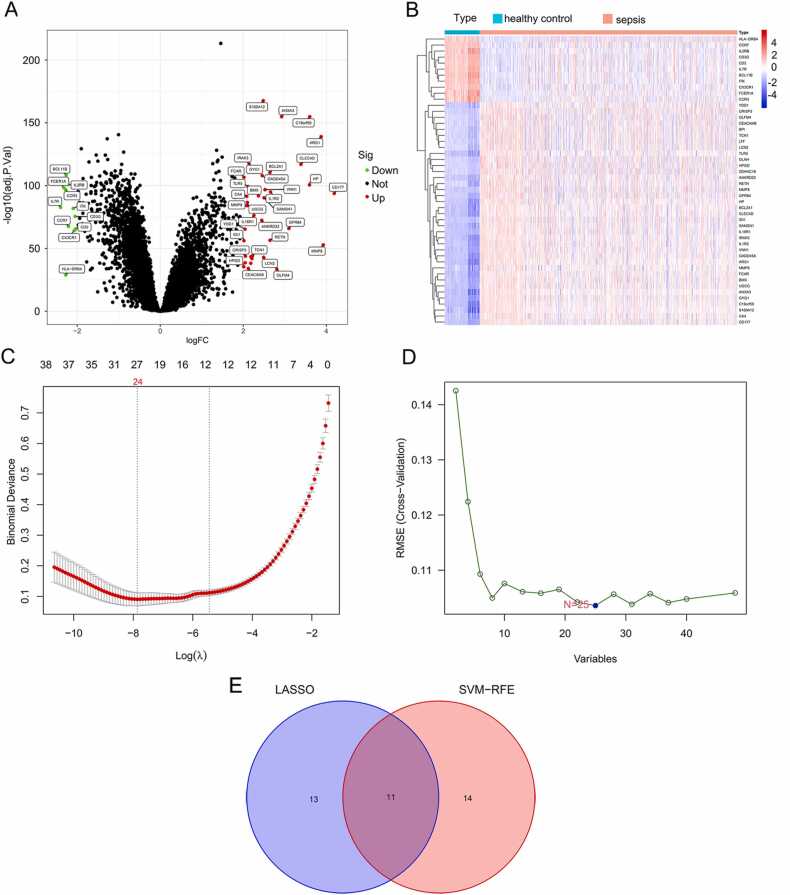
Fig. 1.
Identification of DEGs in sepsis. (A) Volcano map of all DEGs in sepsis and healthy control groups. Red plots represent up-regulated mRNAs with P < 0.05 and log2FC > 2. Green plots represent down-regulated genes with P < 0.05 and log2FC < −2. Black plots represent normally expressed mRNAs. (B) Heatmap of all DEGs. The horizontal axis represents the sample, and the vertical axis represents different genes; the red color indicates increased gene expression, and the blue indicates decreased gene expression. (C) λ 1 standard error (λ.1 SE) usually optimizes regularization so that the error and minimum error remain within a standard deviation error. Optimal λ value identified by using 10-fold cross-validation via minimum and 1-SE criteria in the LASSO regression analysis. Two marked dashed lines indicate two special lambda values λ. min and λ.1SE, and the λ between the two values is considered appropriate. λ. 1SE builds the simplest model by using fewer genes. λ. min was more accurate with using a larger number of genes. (D) SVM-RFE algorithm. The horizontal axis represents the number of DEG variables. The vertical axis represents cross-validation RMSEs. The marked plot is the number of DEGs required to obtain the optimal value. (E) Venn diagram of overlapping genes selected by Lasso and SVM-RFE algorithms.