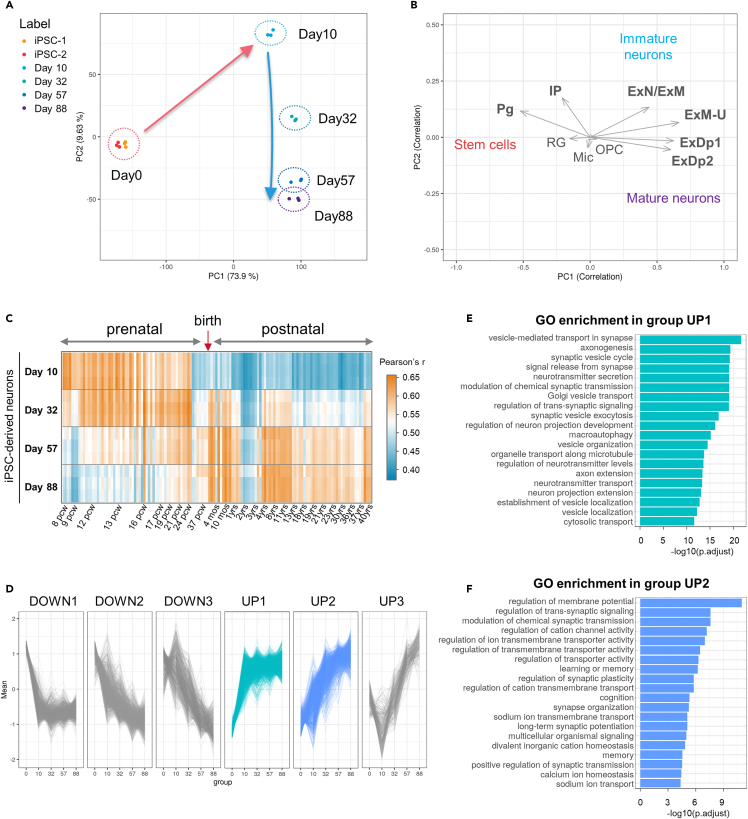
Figure 2.
Global transcriptome analysis of cultured TF-induced iPSC neurons by RNA-Seq
(A) Principal component analysis (PCA) of the cell samples at day 0 (iPSC-1 and iPSC-2), days 10, 32, 57, and 88 of culture following the induction of differentiation.
(B) Correlation with the factor loadings of various brain cell types of the human neocortex. The direction most correlated with a specific cell type is represented as the average factor loading vector of the cell type-specific marker genes. The cell types have been selected and annotated according to the definitions from the CoDEx dataset.27
(C) A correlation heatmap of the gene expression in TF-induced iPSC neurons, compared with the developing human brain of the Brainspan dataset.28 The analysis includes data from the prefrontal cortex in the prenatal phase (post-conception week 8-26) and the postnatal/adult phase (4 months-40 years following birth).
(D) Gene expression profiles categorized into six patterns by hierarchical clustering. Each line is plotted from the mean expression levels of a gene with a significant variation across the time-course.
(E and F) The leading 20 significantly enriched Gene Ontology (GO) terms for biological processes in groups UP1 and UP2. The GO enrichment analyses for other groups are reported in Figure S4.
Pg: Cycling progenitors; IP: Intermediate progenitors; ExN: Migrating excitatory neurons; ExM: Maturing excitatory neurons; ExM-U: Maturing excitatory upper enriched; ExDp1: Excitatory deep layer 1; ExDp2: Excitatory deep layer 2; RG: Radial Glia; Mic: Microglia; and OPC: Oligodendrocyte precursor cells.