Figure 1.
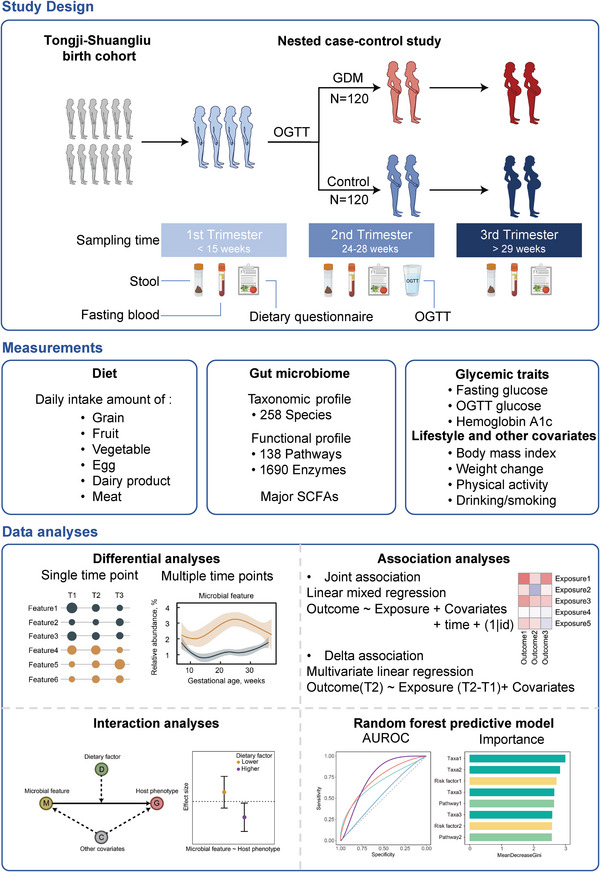
Study design, measurements, and analysis strategy. To associate the gut microbiome with diet and glucose metabolism, we profiled stool metagenomes and glycemic traits from a prospective case‐control study nested in the Tongji‐Shuangliu birth cohort. Blood and stool samples, dietary records, and health‐related information were collected, and taxonomic and functional profiling from stool shotgun metagenomes, fecal short chain fatty acids (SCFAs), plasma biomarkers of glucose metabolism, and other covariates were measured at each trimester (n = 720). The joint associations between microbial features and glycemic traits were estimated using linear mixed models with pooled data from three trimesters (n = 720). The delta associations were estimated using the change values of microbial features between the first trimester (T1) and the second trimester (T2) and oral glucose tolerance test (OGTT) measured in T2 (n = 240). Interaction analyses were performed to explore the potential interaction effect of dietary factors on the associations between microbial features and host glucose metabolism. Random forest classification models were constructed to estimate the prediction power of microbial data for the risk of gestational diabetes mellitus (GDM). The area under operating characteristic curve (AUROC) was used as a metric to quantify classifiers performance. Icons representing the types of collected samples were created using Biorender.com.
