Figure 6.
Excess of dgRNA reduced KI efficiency and drastically increased on-target large deletion in a point mutation rat animal model
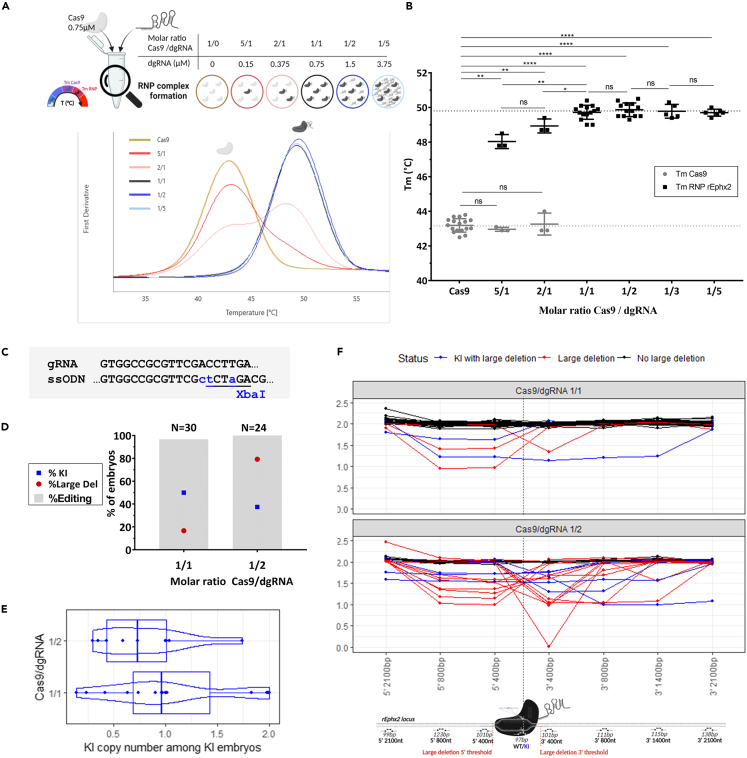
(A) Schematic representation of nanoDSF assay and associated derivative curves used to determine Tm of Cas9 (TmCas9) and RNP complex (TmRNP) for each concentration of dgRNA rEphx2 with a constant concentration of Cas9 (0.75 μM).
(B) Comparison of RNP complex formation efficiency with increasing concentrations of dgRNA in the same conditions described in A. by analysis of TmCas9 (gray) and TmRNP (black) with dgRNA rEphx2. Data are represented as individual replicates with mean ± SEM. Asterisks highlight the statistical differences between conditions using unpaired nonparametric Mann-Whitney t-tests (p > 0.05 (ns), ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001).
(C) Schematic representation of the point mutation generated by KI with a XbaI ssODN on the rat Ephx2 locus.
(D) Comparison of KI (blue squares), total editing (gray bars) and large deletion (red dots) rates with an equimolar (1/1) Cas9/dgRNA ratio or excess of dgRNA (1/2) and a constant dose of Cas9 (0.2 μM) and ssODN (2 μM). Conditions with Cas9/dgRNA ratio 1/1 (N = 30) and 1/2 (N = 24) were electroporated on 3 different days.
(E) Distribution of KI copy number among KI embryos with the same conditions as described in D. using a boxplot combined with a violin plot. Points represent each embryo carrying KI allele. No significant difference was noted when Kruskal-Wallis test was done.
(F) Allele copy number analysis around Cas9 cleavage site for each condition by ddPCR on rat embryos presenting no large deletion (black), large deletions with (blue) or without KI allele (red). Each curve represents one embryo and each point a ddPCR assay. The figures were created with https://www.biorender.com/ (Toronto, Canada). dgRNA, dual guide RNA; RNP, ribonucleoprotein complex; Tm, melting temperature; ns, non-significant, ssODN, single-stranded oligonucleotide; KI, knockin; bp, base pair; nt, nucleotide.