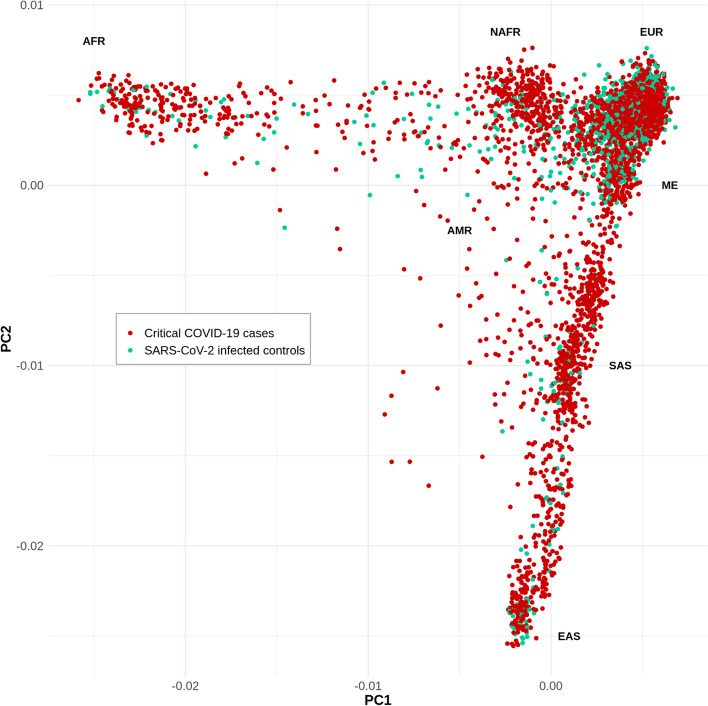
Fig. 2.
Principal component analysis of patients with life-threatening COVID-19 (red) and SARS-CoV-2-infected controls (green). Principal component analysis (PCA) was performed with PLINK v1.9 software [40] on a pruned subset of ~ 14,600 exonic SNPs in linkage equilibrium (maximum r2 value for linkage disequilibrium of 0.4 between pairs of SNPs) with a minor allele frequency (MAF) > 1%, call rate > 99% and P value for departure from Hardy–Weinberg equilibrium > 10.−5. Samples were of diverse ethnic origins, including European (EUR), admixed American (AMR), North African (NAFR), sub-Saharan African (AFR), Middle Eastern (ME), South Asian (SAS), and East Asian (EAS)