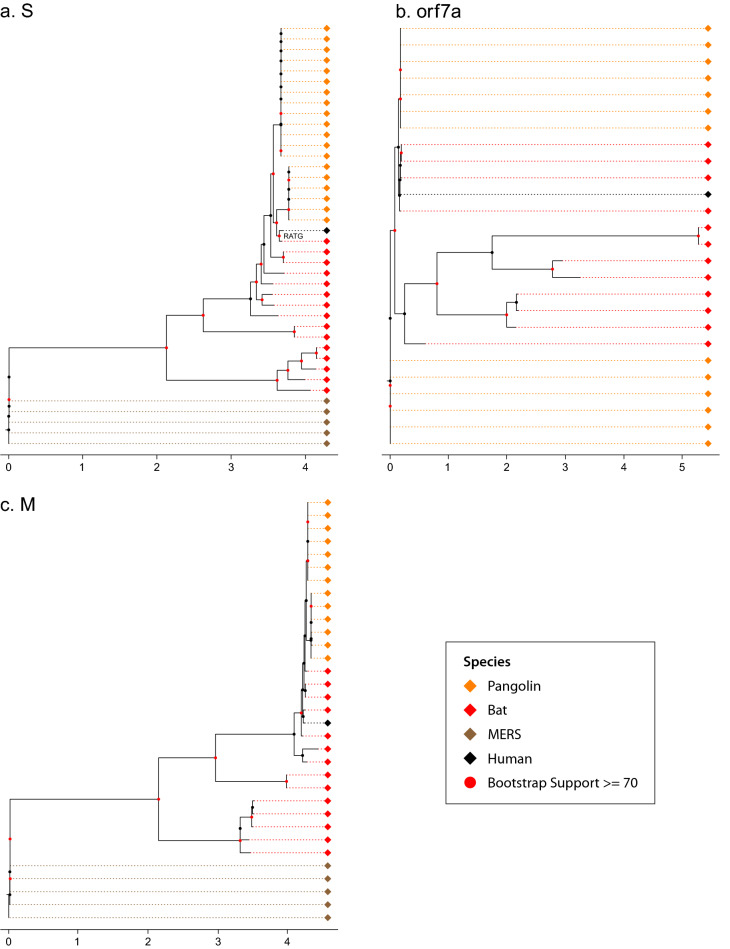
Figure 1.
The consensus gene trees of the S, ORF 7a, and M genes are discordant: (A) S; (B) ORF 7a; (C) M. The gene trees were reconstructed by the method of maximum likelihood19 from 5,248 viral genomes. The consensus gene trees were reconstructed from a bootstrap sample of 100 gene trees. Human, bat, pangolin, and MERS-CoV coronavirus sequences are color-coded as black, pink, orange, and dark orange, respectively. The MERS CoV genomes root the gene trees. All 11 gene trees for the coronavirus genome are in the supplement (Figs. S1–S11). The black diamond represents the amalgamation of human SARS-CoV-2 lineages. A common scale bar for tree branch lengths is provided. The branch lengths on Fig. 1 are the mean of the expected number of substitutions per site across all 100 gene trees in the bootstrap sample of each gene (see Materials and Methods). Red dots on the trees indicate nodes with at least 70% bootstrap support.