Fig. 1.
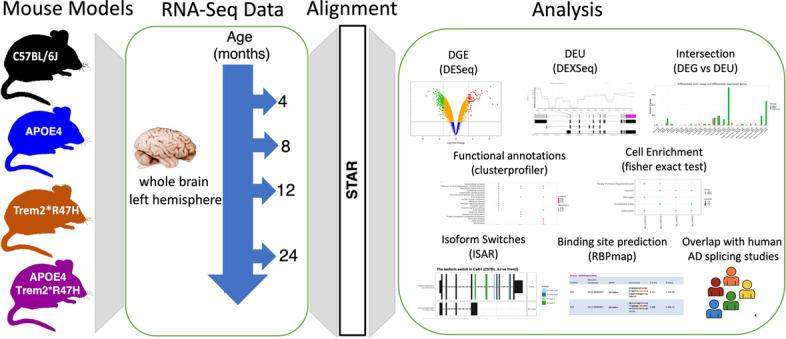
Schematic overview of the analysis. Bulk RNA-Seq data from mouse whole brain hemispheres were processed by our parallelized RNA-Seq analysis pipeline. We performed differential gene expression analysis, differential exon usage analysis, and differential isoform usage analysis. After performing differential analysis, we used the software clusterProfiler to perform functional enrichment analysis followed by cell type enrichment analysis. The RBPmap webserver was used to predict Binding sites of RNA-binding proteins on differentially spliced genes. In addition, we performed hypergeometric tests to identify enrichment of differentially spliced gene set in human Alzheimer’s cases
