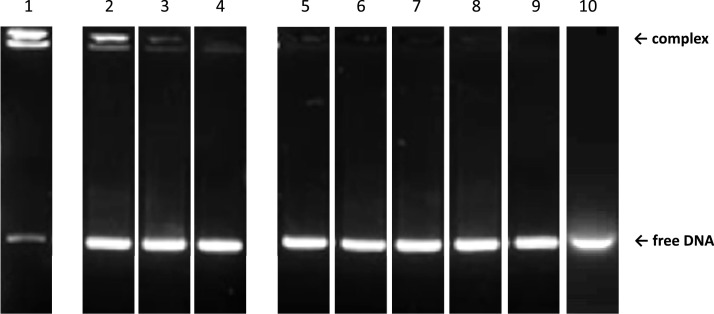
Fig. 1.
EMSA analysis of Hfq-CTR wild type (WT) and mutants in the presence of DNA. Lane 1: Control, Hfq-CTR WT (this control was reprinted with permission from Biomacromolecules 2020, 21, 3668–3677. Copyright 2020 American Chemical Society); lane 2: Hfq-CTR S87A,S88A; lane 3: Hfq-CTR E99A,E100A,E102A; lane 4: Hfq-CTR Y83A. In these case a complex is formed with the peptide. Note that the complex, when it forms, migrates on the top of the gel (but not in the well that is not visible here), indicating that numerous peptides are bound to DNA. Sometimes 2 complexes of different size are present, probably corresponding to different numbers of CTRs bound to DNA [43]. Taking into account the bridging properties of the CTR [38], we suspect these two bands could be (CTRn:AT59) and (CTRn:AT59)2, where 2 (CTRn:AT59) are bridged. Lane 5: Hfq-CTR S80A,S81A; Lane 6: Hfq-CTR R66A; Lane 7: Hfq-CTR S65A,S69A,S72A; Lane 8: Hfq-CTR S93A,S98A; Lane 9: Hfq-CTR H70A,H71A,H84A,H85A; Lane 10: Hfq-CTR G76A, G77A, G78A. In these cases no complex is formed.