Highlights
-
•
Anionic DMPG vesicles promote FapC fibrillation below and above the lipid tm.
-
•
Zwitterionic (DMPC, DOPC) have no effect on FapC fibrillation while DOPG did not show a clear effect.
-
•
Heating of FapC induces higher β-sheet content and rapid fibrillation.
-
•
FapC imperfect repeat 2 is important for reversibility of thermal changes.
-
•
EGCG inhibits FapC-DMPG contacts and redirects FapC to off-pathway oligomers.
Keywords: bacterial amyloid, FapC protein, membrane vesicle, EGCG, ThT kinetics
Abstract
Functional amyloids (FA) are proteins which are evolutionarily optimized to form highly stable fibrillar structures that strengthen the bacterial biofilm matrix. FA such as CsgA (E. coli) and FapC (Pseudomonas) are secreted to the bacterial surface where they integrate into growing fibril structures projecting from the outer membrane. FA are exposed to membrane surfaces in this process, but it remains unclear how membranes can interact with FA and potentially affect the self-assembly. Here we report the effect of different vesicles (DOPG, DMPG, DOPS, DOPC and DMPC) on the kinetics and structural endpoints of FapC fibrillation using various biophysical techniques. Particularly anionic lipids such as DMPG trigger FapC fibrillation, and the protein's second repeat sequence (R2) appears to be important for this interaction. Vesicles formed from phospholipids extracted from three different Pseudomonas strains (Δfap, ΔFapC and pfap) induce FapC fibrillation by accelerating nucleation. The general aggregation inhibitor epigallocatechin gallate (EGCG) inhibits FapC fibrillation by blocking interactions between FapC and vesicles and redirecting FapC monomers to oligomer structures. Our work indicates that biological membranes can contribute significantly to the fibrillation of functional amyloid.
Graphical abstract
Introduction
Amyloid fibrils are highly ordered and β-sheet-rich protein self-assemblies, commonly associated with neurodegenerative diseases such as Alzheimer's and Parkinson's (PD) [1,2]. In addition, they occur in natural contexts as beneficial agents. For example, functional amyloid promotes bacterial biofilm formation by mechanically strengthening the biofilm matrix [3] and promote bacterial resistance to different environmental stresses and antibiotics [4]. Furthermore, they can act as reservoirs for quorum-sensing molecules [5]. In Pseudomonas strains, the protein FapC is the major amyloid-forming component, containing three amyloidogenic conserved imperfect repeats (R1, R2 and R3) connected by two linker regions (L1 and L2) of variable length [6], [7], [8]. Mature FapC from Pseudomonas sp. UK4 (without the 24-residue signaling peptid) consists of 226 amino acids with an isoelectric point (pI) 6.91, net charge of -1.5 at pH 7.4 and a mass of 22.56 kDa without considering the first 24-residue signaling peptide (see FapC sequence provided in SI). FapC is produced from a six-gene operon known as fap (functional amyloid in Pseudomonas) consisting of fapABCDEF. Besides FapC, the other components play various roles in amyloid biogenesis. FapA is likely a chaperone which keeps FapC unfolded in the periplasmic space before it is secreted through the outer membrane via the membrane pore FapF [8]. FapB is likely a nucleator attached to the bacterial surface that triggers FapC fibrillation [6]. FapE may help transport FapC through FapF via contacts between C-terminal Cys residues in FapE and FapC. Finally FapD is likely a cysteine protease that processes several Fap proteins [9].
Amyloid formation is often highly sensitive to environmental conditions. Biomolecules such as biosurfactants [10], lipopolysaccharides (LPS) [10], heparin [11], fibrinogen [12] and chaperones [13] can induce or inhibit fibrillation. During transport over the periplasmic space and after export from the outer membrane, FapC has multiple opportunities to interact with membrane surfaces. Lipid properties such as charge, headgroup composition and ionic strength affect these interactions and change various aspects of the fibrillation process. For example, the Aβ peptide forms short fibrils in the presence of DOPC vesicles but longer fibrils with vesicles containing negatively charged headgroups like DOPG and DOPS [14]. Of equal importance, these membranes greatly accelerate fibrillation of Aβ and both the fibrillation process and formation of toxic oligomers can lead to membrane disruption [15,16]. Aggregation of the peptide IAPP is also strongly promoted by anionic lipid membranes [17] whereas zwitterionic phospholipids like DOPC have no effect [18]. High membrane curvature (for vesicles of 30 nm of less) lead to more packing defects that may encourage intractions between protein and membrane [19]. For example, it promotes binding, conformational changes and fibrillation of Aβ via nucleation of monomer on the vesicle surface [20]. SUV membrane studies are particularly pertinent for neurodegenerative diseases involving small synaptic vesicles, whereas the bacterial membrane surface (with a cell size of ∼1 µm) ise more appropriately mimicked by LUVs. Other lipid-dependent factors of importance for aggregation include size and shape of lipid head groups and the hydrophobic tail as well as membrane fluidity [21]. The 140-residue protein α-synuclein (α-syn), strongly involved in the development of Parkinson's Disease, is known to interact strongly with membranes (particularly anionic lipids). PC and PG lipids respectively inhibit and stimulate α-syn fibril formation; the N-terminal Lys-rich region of α-syn interacts strongly with the negatively charged PG, leading to a marked acceleration of fibrillation kinetics [22,23]. Aggregation of α-syn in the presence of membranes leads to complete membrane disruption, while α-syn oligomers are associated with membrane permeabilization, cell death and Lewy Body formation [24]. These intimate protein-membrane interactions prompted us to investigate how phospholipid vesicles affect fibrillation of FapC. The two major lipid headgroup in Gram positive bacteria are both anionic, namely PG and cardiolipin (CL) which in essence is two PG moieties connected by a glycerol backbone [25]. These lipids are accompanied by PE and PI lipids as well as O-aminoacyl derivates of PG. Gram negative bacteria contain an additional outer membrane (OM) that is stabilized by lipopolysaccharides (LPS) in the outer leaflet. The OM contains variable amounts of PC, PE, CL and PG phospholipids in different bacteria [26]. We previously reported that LPS trigger FapC fibrillation by decreasing the lag time of nucleation phase [10]. In this study, we focused on the effect of lipid membrane on FapC fibrillation. For this purpose, we selected four different vesicles containing lipids with different phase transition temperatures that usually are present in bacterial membrane (DOPC, DOPG, DMPC and DMPG with melting temperatures of -17, -18, 24 and 23 °C, respectively).
Based on a range of biophysical analyses, we conclude that the anionic DMPG induces fibrillation, both in the gel phase and in the liquid disordered phase (i.e. below and above its melting temperature tm) while zwitterionic lipids (DMPC and DOPC) and liquid-disordered anionic lipids (DOPG and DOPS) have no significant effect. Membrane extracts obtained from three different Pseudomonas strains all promote FapC fibrillation by accelerating nucleation. Thus membrane lipids can have a varied impact on FapC fibrillation.
Materials and methods
Materials
1, 2-dioleoyl-sn-3-phosphatidylglycerol (DOPG), 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DMPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) were from Avanti Polar Lipids (Alabaster, AL). Other chemicals were from Sigma Aldrich (St. Louis, MO) at highest purity grade. Recombinant His-tagged FapC and 4 FapC mutants (ΔR1R2, ΔR1R3, ΔR2R3 and ΔR1R2R3) were purified as described [10]. Subsequently proteins were stored in 8 M guanidinium chloride at -20°C. Immediately prior to use, the protein solution was desalted with a PD-10 column into PBS buffer pH 7.5. The sequence of full-length FapC highlighting the individual repeats is shown in Supplementary Information Fig. S1.
Preparation of vesicles (Large Unilamellar Vesicles (LUV)
5 mg/ml of four different lipids (DOPG, DMPG, DMPC or DOPC) were dissolved in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4). and subjected to 10 freeze-thaw cycles between liquid nitrogen and a 50°C water bath followed by 21 rounds of extrusion using an Avanti extruder and a 100 nm filter. DMPG vesicles were extruded at 50°C using a hot plate to stay above the tm (23°C). Vesicles were used within a few h of preparation.
Purification of membrane phospholipids (MPL) with Membrane Proteins (MP)
Pseudomonas bacteria were grown overnight on Luria-Bertani (LB)-tetracycline (30µg/ml) agar plates at 28°C. One colony was resuspended in 1 ml LB medium and streaked out on four new LB-agar plate with tetracycline and incubated overnight at 28°C. 0.5 ml of IPTG (0.5 mM) was added to each plate and incubated until the next day. The colonies were washed off and resuspended in 50 mM Tris pH 8 containing 1 mM PMSF (0.1 mM). The bacteria cells were homogenized using a homogenizer (Potter-Elvehjem). 0.5 ml of enzymes containing 2 mg/mL RNase, 2 mg/mL DNase I, 4 mg/mL lysozyme, 20 mM MgCl2 were added to the sample and subsequently subjected to 2 cycles of French Press (Homogenizers, Atkinson, NH) to lyse and disrupt bacterial cells by shear force. Cell debris was removed by centrifuging the sample at 15000 × g for 30 min, and membrane phospholipids and membrane proteins were purified by centrifuging the supernatant at 30,000 rpm for 1 h at 4 °C. The pellet was collected and resuspended in 15 ml of 50 mM Tris pH 8 containing PMSF.
FapC fibrillation assays
30 µM FapC was incubated with 30–1500 µM (monomer lipid equivalents) of vesicles in PBS at 37°C with 40 µM ThT in final volume of 150 µl in a 96-well plate (Nunc, Thermo Fisher Scientific, Roskilde, DK) covered with clear sealing tape (Hampton Research, Aliso Viejo, CA) to prevent evaporation. The plate was shaken for 5 s every 5 min and ThT-emission measured over a 70-h period using excitation and emission at 448 nm and 485 nm respectively on a Clariostar plus plate reader (BMG Labtech, Ortenberg, Germany). For experiments with Pseudomonas membrane phospholipids, 20 µM of FapC was incubated with different dilutions of the obtained stock solution of purified membrane extracts. The plate was incubated as described above and subjected to 5 s shaking every 10 min. All fibrillation measurements were carried out in triplicate.
SDS-PAGE to measure level of monomeric protein
FapC incubated in the presence and absence of DMPG as described above were centrifuged for 15 min at 13000 rpm in a bench-top centrifuge. 20 µl of supernatant was mixed with SDS-PAGE sample loading buffer and boiled for 5 min, after which10 µl was loaded on a 15% polyacrylamide gel together with a Mark12 molecular weight standard (ThermoFisher, Waltham, MA).
Circular dichroism kinetic and thermal measurements
CD measurements over time were recorded on a Chirascan CD spectrophotometer (Applied Photophysics, Surrey, UK). 15µM of FapC and mutants were incubated with/without 300 µM of DMPG vesicles in a 0.1 mm quartz crystal cuvette with a probe inserted into the cuvette to monitor and control the temperature of the sample. CD spectra were recorded every 10 min. Thermal scans were carried out with the same concentrations of FapC and vesicles. The start and end point of measurements was set to 5 and 90°C and the samples were kept on ice before mixing them, and the curve recording started immediately after mixing FapC and vesicles. The measurements were done in 5°C intervals with stepped ramping selected to two minutes, giving the sample 2 min time to reach the new temperature. After reaching 90°C, the reverse sequence from 90 to 5 °C was then carried out. Each CD spectrum was recorded from 200 to 260 nm in steps of 1 nm. Using the BeStSel web server, thermal CD spectra were deconvoluted by using the “multiple spectra analysis” option to clarify the structural change during incubation with vesicles [27]. Note that we did not see visible precipitation of our FapC samples over time, indicating that light scattering was not significant.
Size exclusion chromatography (SEC) and Small-angle X-ray scattering (SAXS)
30 µM of FapC was incubated with 120 µM EGCG and 200 µM DMPG in the plate reader as described above. After the ThT signal reached a plateau, 1 ml of sample was spun down at 13,000 rpm for 10 min and supernatant loaded on a 24 ml Superose 6 10/300 column (GE Healthcare) at 0.5 ml/ min flow rate. UV absorbance was then measured at 280 nm. Fractions containing oligomers were combined and analyzed on an optimized NanoSTAR SAXS from Bruker AXS as described [10].
Transmission electron microscopy (TEM)
Samples were stained with phosphotungstic acid, and images were recorded on a JEM 1010 electron microscope as described [28].
Results
Different lipids have different effects on FapC fibrillation
To investigate the effect of membranes phospholipids on FapC fibrillation, four different vesicles consisting of two zwitterionic phospholipids (DMPC and DOPC) and two negatively charged phospholipids (DOPG and DMPG) were incubated with FapC and fibrillation was followed by ThT. While DOPC and DMPC do not affect FapC fibrillation, the situation is more complex for anionic lipids; DMPG promotes fibrillation kinetics while DOPG slightly inhibits it (normalized data and summary shown in Fig. 1; raw data in Fig. S2). To elucidate which part(s) of the aggregation process were mainly affected by DOPG and DMPG, the kinetic time courses of fibrillation of FapC in the presence of the vesicles were fitted globally using the webserver programme Amylofit [29] using various aggregation models. The smallest mean residual error (MRE) was obtained using a nucleation elongation model involving rate constants for nucleation (kn) and elongation (k+) and a nucleus of size nc (results summarized in Table 2). In our fitting we allowed the compound parameter k+kn to vary with vesicle concentration while nc was constrained to be the same for all time curves with a given vesicle type. DMPG vesicles increased the compound rate constant, k+kn from 1.95 × 10+3 to 1.66 × 10+4 conc−nc time−2 between 0 and 1.5 mM DMPG. The situation was more complex for DOPG vesicles, which slightly decreased k+kn between 0 and 300 µM DOPG, followed by an increase. These changes are also reflected in the t½ values, which show an abrupt decrease in the presence of DMPG followed by a relatively stable value, whereas DOPG leads to an increase in t½ followed by a decrease (Fig. 1E). In line with the lack of impact of DOPC and DMPC on FapC fibrillation kinetics, it was sufficient for a satisfactory fit to maintain the same value for k+kn in DOPC and DMPC as in the absence of vesicles (Table 2).
Fig. 1.
ThT time profiles for amyloid formation of 30 µM FapC in presence of 0–1500 µM (monomer units) of different lipids. (A) DOPC vesicles (B) DMPC vesicles (C) DOPG vesicles and (D) DMPG vesicles. (E) Plot of t½ versus different lipid concentration obtained from graphs A-D. All measurements were in triplicate and three repeats are shown. (F) Chemical structure of phospholipids used in this study.
Table 2.
Kinetic parameters for aggregation of FapC alone and in the presence of different vesiclesa.
| Vesicle conc. (µM) | DOPC | DOPG | DMPG | DMPC |
|---|---|---|---|---|
| Variable parameter | ||||
| k+kn (conc−nctime−2) | ||||
| 0 | 1,950b | 1,950 | 1,950b | |
| 30 | 1,070 | 8,000 | ||
| 90 | 803 | 8,380 | ||
| 180 | 972 | 9,420 | ||
| 300 | 992 | 10,100 | ||
| 600 | 1,250 | 11,900 | ||
| 900 | 1,640 | 13,500 | ||
| 1500 | 1,870 | 16,600 | ||
| MRE | 0.00106 | 0.00096 | 0.00110 | 0.00106 |
| Globally fixed parametersc | ||||
| nc | 1.17 | |||
Data obtained from fits of a nucleation-elongation model to ThT fibrillation time curves (Fig. 1) using Amylofit. MRE <0.002
No significant variation in kinetics in the presence of vesicles.
Values constrained to a single value at all vesicle concentrations.
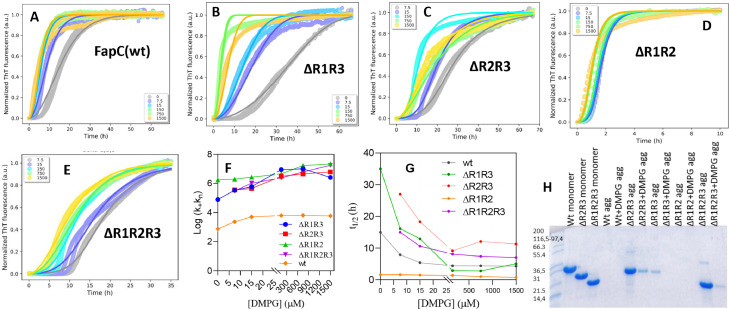
Repeat 2 (ΔR1R3) in FapC protein are more affected by DMPG
To investigate the effect of the most efficacious lipid DMPG in more detail, we studied its impact on the fibrillation of four different FapC mutants in which either 2 or 3 of the protein's 3 imperfect repeats have been deleted (previously analyzed in the absence of vesicles in [7]). The fitted data are shown in Fig. 2, raw data in Fig. S3. In the absence of DMPG, two mutants ΔR2R3 and ΔR1R2R3 did not fibrillate on their own during the 68 h observation window, but their fibrillation was accelerated by DMPG. Aggregation of ΔR1R3, which fibrillates slowly on its own, was strongly accelerated by DMPG (Fig. 2B). In contrast, fibrillation of mutant ΔR1R2 was only marginally affected by DMPG, most likely due to its very rapid aggregation kinetics (Fig. 2D).
Fig. 2.
ThT time profiles for 30 µM truncated FapC mutants incubated with 0-1500 µM (monomer units) DMPG vesicles. Data for FapC wild type are reproduced from Fig. 1A for ease of comparison. All measurements were in triplicate and three repeats are shown. Raw data from panels A-E are used to obtain plots of (F) log k+kn and (G) t½versus [DMPG] for different FapC mutants. (H) SDS-PAGE of supernatants after centrifuging solutions of FapC and different mutants that had previously been incubated with 150 µM DMPG vesicles.
In contrast to fitting of wt data, the ThT time curves for the 4 mutants were best fitted with a model involving secondary nucleation with associated rate constant k2 and nucleation parameter n2. Here k+k2, nc and n2 were globally fixed and only k+kn was allowed to vary with DMPG concentration. For all 4 mutants, k+kn increased with DMPG (Fig. 2F, Table S1). Since the other compound parameter k+k2 could be kept constant during fitting, we conclude that kn (rather than k+) is the parameter mainly affected by DMPG. This likely occurs as a consequence of DMPG increasing the local FapC concentration on the vesicle surface, leading to a reduction in the time of nucleation. The half times of fibrillation (Fig. 2G) decreased significantly for all mutants apart from ΔR1R2, which aggregated so rapidly that vesicles did not make much of a difference. We used centrifugation and SDS-PAGE to probe how much soluble protein was left after incubation of FapC mutants with and without DMPG vesicles (Fig. 2H). The thick bands for DMPG-free ΔR2R3 and ΔR1R2R3 supernatant samples indicate that most of the protein did not aggregate to fibrils in the absence of DMPG (consistent with the lack of ThT signals in Fig. 2CE). However, addition of DMPG led to the almost complete disappearance of these bands, confirming their transformation to aggregates. FapC wt and ΔR1R3 show no monomer bands when incubated with DMPG vesicles, indicating complete aggregation.
DMPG in gel phase accelerates FapC fibrillation more than in the fluid phase
To address how the different phases of DMPG (which undergoes a phase transition from gel to fluid at tm = 23°C) affect fibrillation of FapC, we used CD to determine the secondary structure of FapC after incubation below and above tm (Fig. 3). At 15°C (DMPG in gel phase) the CD of FapC incubated alone did not show any change from the initial random coil structure over a 48 h incubation, whereas addition of DMPG led to complete fibrillation in same time period, seen by the appearance of a minimum around 218 nm (Fig. 3AB). At 37 °C (DMPG in fluid phase), FapC underwent structural changes both with and without DMPG (Fig. 3CD), but as monitored by the change in ellipticity at 218 nm over time (Fig. 3E) as well as the overall content of β-sheet obtained by spectral deconvolution (Fig. 3F), there was a more extensive transition in the presence of DMPG. These results demonstrate that vesicles at both gel and fluid phase can accelerate fibrillation of FapC but – rather counterintuitively - that the gel phase is more effective. Deconvolutions of kinetic CD spectra also indicate the β-sheet content of the FapC incubated with DMPG at 15°C reaches 35%, which is ∼10% more than at 37 °C (Fig. 3F). Remarkably, the anionic vesicle DOPG, which is in the fluid phase under all experimental conditions (tm = -18 °C), did not lead to any significant fibrillation during 60 hours of incubation (Fig. 3G).
Fig. 3.
Changes in secondary structure of 15 µM FapC wt during incubation in presence and absence of 300 µM lipids monitored by far-UV CD. Panels A-D and G provide far-UV CD spectra of FapC with and without either DMPG or DOPG at the indicated temperatures over a 20-60 hr period. Arrows indicate the direction of time. (E) Time course of the development in ellipticity at 218 nm using spectral data from panels A-D. (F) FapC parallel β-sheet content obtained from deconvolution of data from curves A-D using BeStSel [27].
Conformational change in the presence of DMPG at different temperature
To explore the effect of phase changes and temperature on fibrillation of FapC, we heated up FapC wt and different FapC truncation mutants from 5°C to 90°C in the presence and absence of DMPG while recording far-UV CD spectra (Fig. S4). The spectra were subsequently deconvoluted to provide relative fractions of different secondary structures, including both parallel and antiparallel β-sheets along with α-helices (Fig. 4). Each temperature scan (5°C to 90°C) lasted around 45 min. To minimize structural changes before the thermal scan, the samples were desalted and kept on ice until the experiment started. Although the content of organized secondary structure was remarkably high for an IDP such as FapC, we note that the deconvolution fits were generally of high quality with low normalized root mean square deviation. Further, the presence of some β-sheet structure from the early stages of incubation are consistent with FapC's inherent tendency to form cross-β amyloid structures.
Fig. 4.
Deconvolution of the far-UV CD spectra of FapC and mutants from Fig. 3 incubated without (top row) and with (middle row) DMPG. Deconvolution was done using BeStSel [20]. Bottom row shows CD data at 203 nm for wt, ΔR1R3, ΔR2R3 and ΔR1R2R3 respectively during heating up/cooling down in presence and absence of DMPG to test for reversibility of structural changes.
FapC wt starts to accommulate α-helical structure from 30°C onwards (i.e. in the liquid phase of DMPG) (Fig. 4A and B). The structural change occurs both with and without DMPG, but DMPG induces higher levels of helicity. The truncated variants generally showed a lower tendency to aggregate with temperature than wt. In the absence of DMPG, only FapCΔR1R3 led to a modest (10%) increase in helicity and corresponding loss of β-sheet content and this only occurred above 50°C. DMPG accentuated these tendencies and led to a 40% increase in helicity for FapCΔR1R3 and lower amounts for the triple deletion mutant. We also tested the reversibility of this process by monitoring CD spectra during cooling down at the same scan rate. For ΔR1R3 there was complete recovery of the original lack of structure upon cooling down (Fig. 4F). That is, the α-helix content decreased and the antiparallel β-sheet increased to original state from from 90–5 °C (Fig. S5B). Clearly ΔR1R3 was not trapped irreversibly by the heating process. This indicates that repeat 2 in the FapC sequence (the only repeat left in this construct) did not trap the protein in an irreversible state (Fig. S5A). The other mutants did not show any significant change (Figs. 4I, L and S5), except for ΔR1R2 which fibrillated completely during the experiment according to CD spectra (Fig. S5GH). The secondary structural changes of ΔR1R2 is similar to FapC wt in presence of DMPG, showing an increase in parallel β-sheet but helicity and decrease in anti-parallel β-sheet content (Fig. S5G and H). In addition, both wt (Fig. 4C) and ΔR1R2 (Fig. S6) underwent irreversible changes (i.e. fibrillation) according to CD during heating in the presence of DMPG. Our thermal data shows that DMPG encourages the formation of α-helix structures in FapC protein before fibrillation. Furthermore, repeat 2 seems to play a central role in these structural changes, since only truncation mutants lacking this repeat (ΔR2R3 and ΔR1R2R3) failed to show the significant change in helicity that is seen for FapC wt.
Vesicles of different phospholipids lead to different conformational changes at various temperatures
To clarify the interplay between temperature and vesicle types on FapC fibrillation, we analyzed the secondary structure of FapC in presence of other lipids during heating. These lipids included DMPC (tm = 24 °C), DOPG (tm = -18 °C) and DOPS (tm = -11 °C). Incubation of FapC with DMPC vesicle led to an increase in α-helical content up to 40% until 65°C, after which the helicity decreased with a subsequent increase in parallel β-sheet content (Fig. 5AB). As shown in the previous section, secondary structural change including increase in helicity and decrease in antiparallel β-sheet for FapC in presence of DMPG (tm = 24 °C) start around 25 °C (i.e. close to the lipid's tm), whereas for DMPC these conformational changes begin slowly from 5 °C onwards. The difference in behavior must be attributed to the difference in charge between the two lipids. In contrast, for DOPG vesicle, the structural changes start around 30 °C (i.e. well above the lipid's tm of -18 °C) as an abrupt increase in α-helical content and decrease in antiparallel β-sheet while parallel β-sheet were formed from 50 °C (Fig. 5C, D). In DOPS (tm -11 °C), there are more modest changes in α-helical and antiparallel β-sheet (Fig. 5E, F) but no increase in parallel β-sheet. DOPG and DOPS both have negative charge and are in the liquid disordered phase throughout, so the protein conformational changes clearly occur independent of lipid phase changes.
Fig. 5.
Structural changes during incubation of 15 µM FapC with 300 µM of vesicles formed by the indicated lipids while increasing the temperature from 5 to 90 °C. Top row: Far-UV CD spectra. Arrows indicate direction of temperature increase. Bottom row: Changes in secondary content as a function of temperature based on deconvolution of the top-row spectra using BeStSel [20].
TEM image of aggregates formed by incubation of FapC with DMPG
We used transmission electron microscopy (TEM) to analyze the morphology of aggregates that formed in the presence and absence of DMPG (Fig. 6). DMPG-free FapC formed a network of well separated fibrils which in the presence of DMPG became much denser (Fig. 6A, B). FapCΔR1R3 show a dense network of thin fibrils both with/without vesicles (Fig. 6C, D). We observe circular structures that we interpret to be vesicles around which fibrils appear to form. For FapC ΔR2R3 in the absence of DMPG vesicles, some short length twisted fibrils were observed whereas in the presence of DMPG, the fibrils are very thin around the vesicles (Fig. 6E, F). For ΔR1R2R3 only a small amount of fibril was observed by TEM whereas DMPG vesicles induced fibril formation to clusters of laterally associated fibrils (Fig. 6G, H). Fibril binding to membrane vesicles has also been reported in a computational model of Aβ fibrillation [30].
Fig. 6.
TEM images of aggregates of 30 µM FapC wt and the indicated truncation mutants formed in the absence (top row) and presence (bottom row) of 300 µM DMPG vesicles, the vesicle size in panel I is 90 ± 5.6 nm. Scale bars indicate 100nm, except for panel I (200 nm).
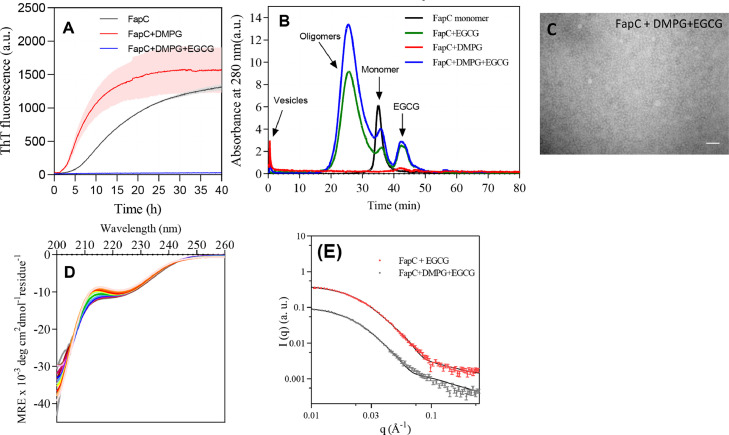
EGCG binds to protein and inhibits the interaction between protein and vesicle
The polyphenol EGCG is known to inhibit FapC aggregation, even in the presence of highly efficient aggregation-inducers such as SDS, LPS and Rhamnolipids [10]. Accordingly, we also investigated the effect of EGCG on FapC fibrillation in presence of DMPG vesicles (Fig. 7A). Unsurprisingly, EGCG completely inhibits FapC fibrillation in the presence of DMPG vesicles. Size exclusion chromatography of FapC shows a high amount of oligomers and an increase in the presence of DMPG vesicles (Fig. 7B). The presence of oligomers was confirmed with TEM (Fig. 7C). Thermal scanning of FapC immediately after mixing with EGCG and DMPG reveals that EGCG suppresses structural changes that FapC would otherwise undergo in its absence (Fig. 7D). Similar experiments done with ΔR1R3 show that EGCG blocks the protein from undergoing the changes in secondary structure that occur in the presence of DMPG vesicles (Fig. 4F), emphasizing that EGCG blocks interactions with DMPG vesicles. Instead EGCG completely redirects FapC to oligomers. Analysis of SAXS scattering curves obtained for these oligomers of FapC and EGCG formed with and without DMPG (Fig. 7E) leads to an average composition of 6 monomers per oligomer (Table 3) in both cases. However, vesicles slightly reduces the fraction of protein found in the disordered shell (Fshell) from 0.39 (no DMPG) to 0.3 (with DMPG), i.e. it modestly increases the fraction of the protein found in the core of oligomer. This can also be concluded from σin (electron density of the core) which is larger for FapC-EGCG-DMPG oligomers than for FapC-EGCG oligomers (Table 3). Finally, the decreased Rg (flexibility of the chains around the core) of FapC-DMPG-EGCG oligomer despite a similar number of monomers per oligomer also suggests that it is more compact than the FapC-EGCG oligomer.
Fig. 7.
Effect of EGCG on FapC fibrillation alone and in the presence of DMPG vesicles and EGCG. (A) ThT time profiles for 30 µM FapC incubated with 200 µM DMPG and 120 µM EGCG, data presented as mean + errors for three repeats. (B) Size exclusion chromatography (SEC) and (C) TEM images of 30 µM FapC aggregated in the presence of 200 µM DMPG with or without 120 µM EGCG. Prior to analysis, samples were incubated in a plate reader until a plateau in ThT fluorescence was reached. Scale bare is 100 nm. (D) CD spectra of of 30 µM FapC in presence of 200 µM DMPG incubated with or without 120 µM EGCG at different temperatures from 5 to 90 °C. Arrow indicates direction of increasing temperatures. (E) Graph of scattering intensity I(q) versus length of scattering q (Å−1) vector of SAXS data from size exclusion chromatography purified oligomers from Panel B.
Table 3.
Properties obtained by analysis of SAXS scattering curves after incubation of 30 µM FapC with 200 µM DMPG vesicles with or without 120 µM EGCG.
| Sample | Na | Fshellb | Rg,chainc (Å) | χ2d | σin (Å) e | σout (Å) f |
|---|---|---|---|---|---|---|
| FapC+EGCG | 6.1 ± 0.1 | 0.39 | 19.5 | 1.5 | 18.2 ± 0.1 | 20.1 ± 0.4 |
| FapC+EGCG+DMPG | 6.0 ± 0.2 | 0.30 | 16.4 | 2.6 | 24.4 ± 8.1 | 22.0 ± 21.8 |
Number of monomers in the oligomer
Mass fraction of protein in the shell (thus mass fraction of protein in the core is 1 - Fshell).
Radius of gyration of the flexible chains that are found in the shell.
A test that compares a model to actual observed data. A value close to 1 suggest a good fit.
Electron density of the core. The radius of the core Rin = 2 σin
Electron density of the shell. The outer radius Rout = 4 σin + 2 σout.
Vesicles of purified membrane phospholipids accelerate FapC fibrillation
Hitherto we have investigated the effects of synthetic phospholipid vesicles. We extended our work to bacterial phospholipids which were purified from three Pseudomonas sp. UK4 strains. Among these, the strains Δfap and ΔFapC lack the genes for the entire fap operon and for FapC respectively, while the Pfap strain overexpresses the FapC protein and the rest of the Fap operon. The membrane fractions we isolate contain other membrane components including LPS, outer membrane lipids and outer membrane proteins for all three strains. Thus in principle the three membrane fractions only differ in that the membrane from the pfap strain has the full Fap machinery, the ΔfapC membranes only lack FapC while Δfap phospholipids lack all Fap proteins though we cannot rule out that the membrane proteome has been altered to a greater extent, as suggested by a proteomic analysis of Pseudomonas with and without overexpression of the fap operon [31].
These phospholipids were extruded to 100 nm vesicles and incubated with FapC while following ThT fluorescence (Figs. 8 and S7). The increased lag phase compared to our previous results reflects a lower FapC concentration (20 versus 30 µM).
Fig. 8.
ThT time profiles from incubation of 20 µM FapC with different dilutions of membrane fractions obtained from Pseudomonas sp. UK4 with the following modifications:(A) Δfap, (B) ΔfapC and (C) pFap. The curves were fitted using Amylofit webserver to provide (D) values for different compound rate constants plotted versus the dilution factor. Plotted data represent the compound rate constant k+kn which is allowed to vary with membrane fraction dilution, while the values of parameters indicated in the boxes are kept fixed for all three membrane fractions.
Incubation of FapC with all three membrane fractions modestly accelerates amyloid fibrillation by reducing the lag phase (Fig. 8). Analysis of these ThT-curves with Amylofit identified secondary nucleation as the model with the lowest MRE. The vesicles induce fibrillation by accelerating nucleation of amyloid formation, shown as an increase in the compound rate constant k+kn (Fig. 8D, Table S2) which occurs to very much the same extent by all 3 membrane fractions. As the rate constant of secondary nucleation k+k2 did not change with increasing vesicles concentration (the value could be kept fixed at a value of 1 × 10+14 for all three membrane fractions), we conclude that the increase in k+kn results from the increase in kn, i.e. the nucleation process.
FapC mostly fibrillates around the vesicles of purified phospholipids
To investigate the architecture of fibril formation in the presence of membrane phospholipid vesicles, we recorded TEM images of the Δfap and ΔfapC vesicles preincubated alone and with monomeric FapC (Fig. 9). While the vesicles from strains lacking the fap operon or FapC themseves do not show any FapC fibrils (Fig. 9A,B), aggregation of FapC in the presence of these fibrils leads to aggregates in association with these vesicles (Fig. 9C,D), which suggests a direct catalytic effect by the vesicles on fibrillation. The width of the fibrils formed with these Δfap vesicles are similar to the FapC-Ctrl fibrils shown in Fig. 9E, i.e. 12.4 ± 0.5 nm. However, in the presence of the ΔFapC strain phospholipids, the thickness of the FapC fibrils decreases to 8.15 ± 0.5 nm width. Intact Pseudomonas cells overexpressing fap show a network of thin-branched fibrils with a width of 8.6 ± 0.5 nm close to the bacterial membrane (Fig. 9F), emphasizing the close association of fibrils with the cell and the clear opportunities for FapC-membrane contacts during biogenesis.
Fig. 9.
TEM images of aggregates formed in presence and absence of vesicles that are formed from membrane fractions of different Pseudomonas sp. UK4 strains. (A) Vesicles from Δfap strain, (B) Vesicles from ΔFapC strain, (C) FapC fibrils formed in presence of Δfap vesicles, (D) FapC fibrils formed in presence of ΔfapC vesicles, (E) FapC fibrils formed without vesicles. (F) Intact cells of Pseudomonas sp. UK4 (pFap) showing bacterial fibrils consisting of FapC (highlighted in dashed box). Scale bars indicate 100 nm.
Discussion
Protein-membrane interactions are important in amyloid self-assembly
Protein-membrane interactions can influence fibrillation of many amyloidogenic protein/peptides [32], [33], [34], [35] and this is usually attributed to the increase in effective (local) concentration of protein, sometimes combined with the induction of amyloidogenic structures as reported for α-syn [36]. Assembly of proteins in amyloid structures has been well studied in case of disease-related amyloid formation but little is known about formation of functional bacterial amyloid in presence of membrane structures. This despite the fact that functional amyloid is formed in close vicinity to the bacterial surface and therefore has good opportunity to interact with membrane components. The composition of the outer membrane is complex and contains LPS as well as phospholipids. We have previously shown that LPS can significantly accelerate FapC aggregation, in good agreement with elegant work by Mukhopadhyay and co-workers on the role of LPS in the aggregation of the functional amyloid curli from E. coli [37]. However, the role of bilayer-forming phospholipids in FapC aggregation has not been investigated. In this study we report that fibrillation of FapC protein in presence of membrane strongly depends on phospholipid composition. Among 5 different membranes (DOPC, DOPG, DOPS, DMPC and DMPG), only DMPG interacts strongly with FapC and induces fibrillation (Fig. 1). Zwitterionic phospholipids did not promote fibrillation. This is consistent with numerous other studies which show that anionic lipids have a much stronger effect on amyloid self-assembly than zwitterionic lipids [33,38]. For example, the functional amyloid TasA also interacts strongly with charged lipid composed of PE/PG/CL [39]. The negatively charged lysolipids (lipids with one hydrophobic tail that form micelle) LPG accelerate fibrillation of FapC around its cmc (critical micelle concentration) via strong interactions with FapC and stabilising intermediates whereas zwitterionic lysolipids LPC only show a weak interaction with FapC [40]. Previously we have shown that positively charged Lys residues have a strong effect in the interaction of Phenol Soluble Modulin (PSM) from S.aureus with negatively charged proteins/molecules like fibrinogen and heparin [11,12]. Similarly, we believe that the 14 Lys residues present in FapC (there are no Arg residues) can promote electrostatic interactions between FapC and negatively charged vesicles.
Heating of FapC can induce amyloid formation
Heating of FapC in buffer leads to a decrease in the amount of antiparallel β-sheet structures in monomeric FapC (particularly from 50°C onwards) and an increase in the amount of parallel β-sheet that correspond to the formation of amyloid fibrils. This fibril formation is irreversible since the original structure is not regained after or during cooling down. In the presence of DMPG, the amyloid formation starts from lower temperature (35°C) but remains irreversible. The remarkable effects of both heating and DMPG suggest that harsh environmental conditions such as elevated temperatures can promote amyloid formation and subsequently strengthen biofilm matrix to protect bacteria under stressful conditions. It is common knowledge that heating induces protein misfolding and amyloid formation of proteins [41,42] but it is interesting to note that higher (but biologically compatible) temperatures led to thicker biofilms [43]. Thus the presence of biological membranes may encourage fibrillation under physiologically relevant conditions to ensure bacterial survival.
Intractions of repeat 2 and 3 play important role in fibrillation of FapC in presence of DMPG
Another important insight is that the effect of vesicles depends on the intrinsic aggregative properties of FapC. Specifically, FapC variants which fibrillate slowly or not at all on their own due to removal of several of the imperfect repeats (ΔR1R3, ΔR1R2 and ΔR1R2R3) are markedly aided in their fibrillation by incubation with DMPG vesicles. This is manifested as a reduction in the nucleation time and probably occurs through an increase in the local concentration of monomer, possibly in combination with a reduction in the intermediate steps in amyloid formation. Thus it seems plausible that the proteins that only aggregate slowly or not at all are greatly aided by DMPG which overcomes the bottleneck of initial intermolecular contact formation; fast fibrillators such as FapC wt and ΔR2R3 are presumably only limited by diffusion and conformational changes associated with fibril growth which are not affected by DMPG.
Unexpectedly, CD spectra of FapC incubated with and without DMPG at a given temperature and measured over time indicate that vesicles in both gel and fluid phases can accelerate fibrillation but the transition in the gel phase is faster, emphasizing the effect of gel phase of vesicles on nucleation step. Typically the fluid phase is a more active medium for promoting membrane-catalyzed processes but there may also be an entropic advantage to the immobilization of proteins on a more ordered surface, which also can provide a more stable and well-defined binding interface for fibril propagation. Nevertheless, the incubation of FapC in presence of DMPG reduces FapC's antiparallel β-sheet content around tm of vesicle and increases the content of α-helix and parallel β-sheet, of which the latter is expected to help fibrillation. It is however possible that this effect is partially cancelled by the formation of α-helix which is not a natural component of amyloid. This induction of α-helical structure has also been observed during interactions of other amyloidogenic monomers such as α-syn, Aβ and IAPP with membranes [44], [45], [46], [47], [48]. Molecular dynamic simulations suggest that hydrophobic interactions between Aβ42 and lipid encourage formation of α-helix that may embed into the membrane and subsequently change to β-sheet structure [49]. The lack of vesicles in the TEM images of wild-type fibrils formed in presence of DMPG (Fig. 6B) indicate that fibrillation of proteins in the presence of anionic membranes can lead to membrane disruption, most likely due to disruptive reorganization of the growing amyloid structure while in contact with the vesicle surface. This has also been reported for IAPP [50]. FapC fibrillation was not affected by the presence of zwitterionic DMPC (Fig. 1). Furthermore, the collapse of FapC's α-helix structure in the presence of DMPC vesicles at high temperatures (Fig. 5A) indicates that this interaction is not as strong as with DMPG. Comparison of the conformational changes in different FapC mutants shows that ΔR1R3 (which only has repeat 2) is strongly influenced by incubation with DMPG, whereas FapC mutants that lack repeat 2 do not undergo the same temperature-dependent structural change. Interestingly the conformational changes of ΔR1R3 under heating could be largely reverted by cooling. In contrast, ΔR1R2 (which only has repeat 3) undergo irreversible aggregation during heating. Clearly repeat 2 and repeat 3 play very different roles in the aggregation of FapC.
Furthermore, there are remarkably different effects of different lipids on FapC thermal behavior. In the presence of DMPC, DOPG and DOPS vesicles, there was a decrease in helicity at high temperature (65 °C and 55 °C for DMPC and DOPG respectively) and a low level of helicity at all temperatures in DOPS, indicating that the α-helix structures are not stable in the presence of these vesicles. In contrast, FapC's helical structure is quite stable up to 90°C in DMPG. Binding to the membrane has been shown to lead to a conformational change to α-helix structure for α-syn and subsequently to β-sheet [51], [52], [53]. Similarly, for IAPP, the presence of DOPC/DOPG vesicles during aggregation leads to a decrease in α-helical and intramolecular β-sheet content, and subsequently an increase in intermolecular β-sheet [54]. The basis for this difference remains unclear, given that both DMPG, DOPG and DOPS are anionic lipids which are in the disordered state for most of the thermal experiments, but it illustrates some unusual properties of the different lipids.
EGCG binds to FapC hot spots, inhibits its interaction with membranes and redirect it to off-pathway oligomers
Incubation of FapC with EGCG in presence of DMPG led to complete inhibiting of fibril formation by redirecting monomer to off-pathway oligomers. The on-pathway oligomers are usually unstable and proceed to fibril formation whereas these EGCG-formed oligomers are stable over longer times and only gradually form large insoluble and presumably less organized structures than amyloid fibrils [55]. The oligomers consist of 6 monomers that follow a core-shell structure as we have previously observed [10]. EGCG prevents the interaction between FapC and DMPG according to far-UV CD spectra, maintaining FapC in a random coil from 5 °C to 90 °C. EGCG has consistently been shown to inhibit FapC fibrillation, both alone and in the presence of fibrillation promoters such as SDS, rhamnolipids or lysolipids [56], and DMPG is no exception to this rule. The inhibitory effect of EGCG might go through interaction with repeat 2 in FapC sequence, as this repeat contains a highly amyloidogenic motif (GVNVAA) that we previously identified through peptide microarrays as a strong binding spot for EGCG [57,58]. One difference compared to the surfactants and lysolipids is that the oligomer formed with EGCG in the presence of DMPG seems to have undergone a certain degree of internal rearrangement in shifting mass from the shell to the core; this is not observed for either rhamnolipid or LPS [56] and once again points to a remarkable feature of DMPG vesicles. Several studies indicate that the presence of lipid vesicles can reduce the inhibitory efficacy of EGCG in disease related proteins like Aβ40 and αSN [59,60]. However, lipids do not prevent EGCG from inhibiting the fibrillation of huntingtin [61] and, as shown here, bacterial FapC protein.
Finally, incubation of FapC with vesicles formed from purified phospholipids from Pseudomonas sp. UK4 also confirm that the vesicles with membrane phospholipids can induce fibrillation by affecting the nucleation phase. All three kinds of vesicles from three different strains Δfap, ΔFapC and Pfap accelerate fibrillation to the same extent, indicating that the phospholipid composition of the membrane extract are more important than the membrane proteins from fap operon. It might have been expected that the presence of FapB in ΔFapC and both FapB/FapC in Pfap strains could promote fibrillation of exogenous FapC compared to Δfap membrane fractions, but there was no obvious difference in ThT fibrillation profiles. Ensuing analysis with Amylofit indicate that the main effect of the vesicles is to accelerate nucleation by increasing the value of the compound parameter k+kn by a factor of 10, which is quite comparable to the effect of increasing concentrations of DMPG vesicles (compare Figs. 2F and 8D).
What are the implications of these lipid effects for FapC's aggregation in vivo? The transport of FapC across the periplasmatic space (where phospholipids are to be accessed via both the inner and the outer membrane) is highly regulated through chaperones which likely block most other interactions (although the likely dynamic nature of these contacts may provide some lipid access). Given how well anionic lipids in particular can stimulate FapC aggregation, this emphasizes the importance of maintaining a highly controlled shunt to the outer membrane. However, once secreted out of the bacteria, the FapC monomers need to translocate to the growing ends of the amyloid fibrils, and during this process it may be possible to access other lipids present in the biofilm. Such lipids might even derive from eukaryotic membranes encountered during biofilm formation in e.g. wounds. Anionic lipids are typically transferred to the outer leaflet of mammalian cells as part of the apoptotic pathway and this could potentially trigger accumulation of amyloid fibrils at such sites, perhaps stimulating new attachment sites for a biofilm colonization.
Conclusion
In this study we report the effect of vesicle bilayers on fibrillation of the functional amyloid protein FapC and show that the anionic lipid DMPG is particularly good at accelerating FapC fibrillation. The interaction of FapC with DMPG likely occurs via repeat 2 in its sequence since the mutant ΔR1R3 is strongly aided in its fibrillation by DMPG and only this mutant's secondary structure was influenced by DMPG vesicles. DMPG show markedly stronger effects on fibrillation than other anionic or zwitterionic lipids and also maintained a more stable α-helical content in FapC than the other lipids. EGCG inhibits the interaction between FapC and DMPG, probably through reacting with the amyloidogenic sequences in repeat 2 and redirecting the FapC monomer to off-pathway oligomers. FapC fibril formation is also promoted by incubation with vesicles consisting of bacterial phospholipids, once again emphasizing the role of the phospholipid membrane in amyloid fibril formation. Table 1
Table 1.
Strains and plasmids.
| Bacteria | Plasmid | Antibiotic | |
|---|---|---|---|
| ΔfapC | Pseudomonas UK4-Δfap-gfp PMMB 190 Tc-UK4fap ΔfapC | pMMB190Tc-UK4fap with a deletion of fapC | Tetracycline |
| Δfap | Pseudomonas UK4-Δfap-gfp PMMB 190 Tc | pMMB190 Δbla tet(TcR) PlacUV5 controlled expression | Tetracycline |
| pfap | Pseudomonas UK4 -Δfap with pMMB190Tc-UK4fap | pMMB190Tc containing the complete UK4 fap operon | Tetracycline |
CRediT authorship contribution statement
Zahra Najarzadeh: Investigation, Visualization, Formal analysis, Writing – original draft. Hossein Mohammad-Beigi: Investigation, Visualization, Formal analysis. Jannik Nedergaard Pedersen: Formal analysis. Gunna Christiansen: Formal analysis. Jan Skov Pedersen: Formal analysis. Janni Nielsen: Formal analysis. Daniel E. Otzen: Investigation, Visualization, Formal analysis, Writing – original draft, Funding acquisition, Project administration, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
D.E.O. is grateful to the Independent Research Foundation Denmark | Natural Sciences (grant no. 8021-00208B and 8021-00133B) for support.
Footnotes
Given his role as Executive Editor, Daniel Erik Otzen had no involvement in the peer-review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to the Associate Editor Maria Laura Mascotti.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2022.100055.
Appendix. Supplementary materials
Data Availability
Data will be made available on request.
References
- 1.Benson M.D., Buxbaum J.N., Eisenberg D.S., Merlini G., Saraiva M.J., Sekijima Y., Sipe J.D., Westermark P. Amyloid nomenclature 2020: update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 2020;27(4):217–222. doi: 10.1080/13506129.2020.1835263. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe K.J., Cyr D.M. Amyloid in neurodegenerative diseases: friend or foe? Semin. Cell Dev. Biol. 2011:476–481. doi: 10.1016/j.semcdb.2011.03.011. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng G., Vad B.S., Dueholm M.S., Christiansen G., Nilsson M., Tolker-Nielsen T., Nielsen P.H., Meyer R.L., Otzen D.E. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 2015;6:1099. doi: 10.3389/fmicb.2015.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Seviour T., Hansen S.H., Yang L., Yau Y.H., Wang V.B., Stenvang M.R., Christiansen G., Marsili E., Givskov M., Chen Y. Functional amyloids keep quorum-sensing molecules in check. J. Biol. Chem. 2015;290(10):6457–6469. doi: 10.1074/jbc.M114.613810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dueholm M.S., Søndergaard M.T., Nilsson M., Christiansen G., Stensballe A., Overgaard M.T., Givskov M., Tolker-Nielsen T., Otzen D.E., Nielsen P.H. Expression of Fap amyloids in P seudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen. 2013;2(3):365–382. doi: 10.1002/mbo3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen C.B., Christiansen G., Vad B.S., Lynggaard C., Enghild J.J., Andreasen M., Otzen D. Imperfect repeats in the functional amyloid protein FapC reduce the tendency to fragment during fibrillation. Protein Sci. 2019;28(3):633–642. doi: 10.1002/pro.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouse S.L., Hawthorne W.J., Berry J.L., Chorev D.S., Ionescu S.A., Lambert S., Stylianou F., Ewert W., Mackie U., Morgan R.M.L. A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis. Nat. Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-00361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perrett S., Buell A.K., Knowles T.P. Springer; 2019. Biological and Bio-inspired Nanomaterials. [Google Scholar]
- 10.Najarzadeh Z., Pedersen J.N., Christiansen G., Shojaosadati S.A., Pedersen J.S., Otzen D.E. Bacterial amphiphiles as amyloid inducers: effect of rhamnolipid and lipopolysaccharide on FapC fibrillation. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2019;1867(11) doi: 10.1016/j.bbapap.2019.140263. [DOI] [PubMed] [Google Scholar]
- 11.Najarzadeh Z., Zaman M., Sereikaite V., Strømgaard K., Andreasen M., Otzen D.E. Heparin promotes fibrillation of most phenol-soluble modulin virulence peptides from Staphylococcus aureus. J. Biol. Chem. 2021;297(2) doi: 10.1016/j.jbc.2021.100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Najarzadeh Z., Nielsen J., Farzadfard A., Sereikaite V., Strømgaard K., Meyer R.L., Otzen D.E. Human fibrinogen inhibits amyloid assembly of most phenol-soluble modulins from staphylococcus aureus. ACS Omega. 2021 doi: 10.1021/acsomega.1c02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagaraj M., Najarzadeh Z., Pansieri J., Biverstål H., Musteikyte G., Smirnovas V., Matthews S., Emanuelsson C., Johansson J., Buxbaum J.N., Morozova-Roche L., Otzen D.E. Chaperones mainly suppress primary nucleation during formation of functional amyloid required for bacterial biofilm formation. Chem. Sci. 2022;13(2):536–553. doi: 10.1039/d1sc05790a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo C.E., Park C.R., Kim H.I. Effect of packing density of lipid vesicles on the Aβ42 fibril polymorphism. Chem. Phys. Lipids. 2021;236 doi: 10.1016/j.chemphyslip.2021.105073. [DOI] [PubMed] [Google Scholar]
- 15.Kotler S.A., Walsh P., Brender J.R., Ramamoorthy A. Differences between amyloid-β aggregation in solution and on the membrane: insights into elucidation of the mechanistic details of Alzheimer's disease. Chem. Soc. Rev. 2014;43(19):6692–6700. doi: 10.1039/c3cs60431d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen P.H., Ramamoorthy A., Sahoo B.R., Zheng J., Faller P., Straub J.E., Dominguez L., Shea J.-E., Dokholyan N.V., De Simone A. Amyloid oligomers: a joint experimental/computational perspective on Alzheimer's disease, Parkinson's disease, type II diabetes, and amyotrophic lateral sclerosis. Chem. Rev. 2021;121(4):2545–2647. doi: 10.1021/acs.chemrev.0c01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayasinghe S.A., Langen R. Membrane interaction of islet amyloid polypeptide. Biochim. Biophys. Acta (BBA)-Biomembr. 2007;1768(8):2002–2009. doi: 10.1016/j.bbamem.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Milardi D., Gazit E., Radford S.E., Xu Y., Gallardo R.U., Caflisch A., Westermark G.T., Westermark P., Rosa C.L., Ramamoorthy A. Proteostasis of islet amyloid polypeptide: A molecular perspective of risk factors and protective strategies for type II diabetes. Chem. Rev. 2021;121(3):1845–1893. doi: 10.1021/acs.chemrev.0c00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terakawa M.S., Lin Y., Kinoshita M., Kanemura S., Itoh D., Sugiki T., Okumura M., Ramamoorthy A., Lee Y.H. Impact of membrane curvature on amyloid aggregation. Biochim. Biophys. Acta (BBA)-Biomembr. 2018;1860(9):1741–1764. doi: 10.1016/j.bbamem.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiura Y., Ikeda K., Nakano M. High membrane curvature enhances binding, conformational changes, and fibrillation of amyloid-β on lipid bilayer surfaces. Langmuir. 2015;31(42):11549–11557. doi: 10.1021/acs.langmuir.5b03332. [DOI] [PubMed] [Google Scholar]
- 21.Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 2012;23(5):886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Kaur U., Lee J.C. Membrane interactions of α-synuclein probed by neutrons and photons. Acc. Chem. Res. 2021;54(2):302–310. doi: 10.1021/acs.accounts.0c00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viennet T., Wördehoff M.M., Uluca B., Poojari C., Shaykhalishahi H., Willbold D., Strodel B., Heise H., Buell A.K., Hoyer W. Structural insights from lipid-bilayer nanodiscs link α-Synuclein membrane-binding modes to amyloid fibril formation. Commun. Biol. 2018;1(1):1–12. doi: 10.1038/s42003-018-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhary H., Stefanovic A.N., Subramaniam V., Claessens M.M. Membrane interactions and fibrillization of α-synuclein play an essential role in membrane disruption. FEBS Lett. 2014;588(23):4457–4463. doi: 10.1016/j.febslet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Goldfine H. Lipids of prokaryotes–structure and distribution. Curr. Top. Membr. Transp. 1982;17:1–43. [Google Scholar]
- 26.Sohlenkamp C., Geiger O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 2016;40(1):133–159. doi: 10.1093/femsre/fuv008. [DOI] [PubMed] [Google Scholar]
- 27.Micsonai A., Wien F., Bulyáki É., Kun J., Moussong É., Lee Y.-H., Goto Y., Réfrégiers M., Kardos J. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46(W1):W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen J.N., Jiang Z., Christiansen G., Lee J.C., Pedersen J.S., Otzen D.E. Lysophospholipids induce fibrillation of the repeat domain of PMEL17 through intermediate core-shell structures. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2019;1867(5):519–528. doi: 10.1016/j.bbapap.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisl G., Kirkegaard J.B., Arosio P., Michaels T.C., Vendruscolo M., Dobson C.M., Linse S., Knowles T.P. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016;11(2):252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 30.Cheng S.Y., Cao Y., Rouzbehani M., Cheng K.H. Coarse-grained MD simulations reveal beta-amyloid fibrils of various sizes bind to interfacial liquid-ordered and liquid-disordered regions in phase separated lipid rafts with diverse membrane-bound conformational states. Biophys. Chem. 2020;260 doi: 10.1016/j.bpc.2020.106355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbst F.A., Sondergaard M.T., Kjeldal H., Stensballe A., Nielsen P.H., Dueholm M.S. Major proteomic changes associated with amyloid-induced biofilm formation in Pseudomonas aeruginosa PAO1. J. Proteome Res. 2015;14(1):72–81. doi: 10.1021/pr500938x. [DOI] [PubMed] [Google Scholar]
- 32.Krausser J., Knowles T.P., Šarić A. Physical mechanisms of amyloid nucleation on fluid membranes. Proc. Natl. Acad. Sci. U. S. A. 2020;117(52):33090–33098. doi: 10.1073/pnas.2007694117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butterfield S.M., Lashuel H.A. Amyloidogenic protein–membrane interactions: mechanistic insight from model systems. Angew. Chem. Int. Ed. 2010;49(33):5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 34.Hellstrand E., Nowacka A., Topgaard D., Linse S., Sparr E. Membrane lipid co-aggregation with α-synuclein fibrils. PLoS One. 2013;8(10):e77235. doi: 10.1371/journal.pone.0077235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.C., Edwald E., Veatch S., Steel D.G., Gafni A. Interactions of amyloid-β peptides on lipid bilayer studied by single molecule imaging and tracking. Biochim. Biophys. Acta (BBA)-Biomembr. 2018;1860(9):1616–1624. doi: 10.1016/j.bbamem.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Galvagnion C., Buell A.K., Meisl G., Michaels T.C., Vendruscolo M., Knowles T.P., Dobson C.M. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat. Chem. Biol. 2015;11(3):229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swasthi H.M., Mukhopadhyay S. Electrostatic lipid-protein interactions sequester the curli amyloid fold on the lipopolysaccharide membrane surface. J. Biol. Chem. 2017;292(48):19861–19872. doi: 10.1074/jbc.M117.815522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds N.P., Soragni A., Rabe M., Verdes D., Liverani E., Handschin S., Riek R., Seeger S. Mechanism of membrane interaction and disruption by α-synuclein. J. Am. Chem. Soc. 2011;133(48):19366–19375. doi: 10.1021/ja2029848. [DOI] [PubMed] [Google Scholar]
- 39.Malishev R., Abbasi R., Jelinek R., Chai L. Bacterial model membranes reshape fibrillation of a functional amyloid protein. Biochemistry. 2018;57(35):5230–5238. doi: 10.1021/acs.biochem.8b00002. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen H.Ø., Otzen D.E., Pedersen J.S. Induction, inhibition, and incorporation: different roles for anionic and zwitterionic lysolipids in the fibrillation of the functional amyloid FapC. J. Biol. Chem. 2022 doi: 10.1016/j.jbc.2022.101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjwal S., Verma S., Röhm K.H., Gursky O. Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 2006;15(3):635–639. doi: 10.1110/ps.051917406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alam M.T., Rizvi A., Rauf M.A., Owais M., Naeem A. Thermal unfolding of human lysozyme induces aggregation: recognition of the aggregates by antisera against the native protein. Int. J. Biol. Macromol. 2018;113:976–982. doi: 10.1016/j.ijbiomac.2018.02.095. [DOI] [PubMed] [Google Scholar]
- 43.Villanueva V.D., Font J., Schwartz T., Romani A.M. Biofilm formation at warming temperature: acceleration of microbial colonization and microbial interactive effects. Biofouling. 2011;27(1):59–71. doi: 10.1080/08927014.2010.538841. [DOI] [PubMed] [Google Scholar]
- 44.Nanga R.P.R., Brender J.R., Vivekanandan S., Ramamoorthy A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim. Biophys. Acta (BBA)-Biomembr. 2011;1808(10):2337–2342. doi: 10.1016/j.bbamem.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skeby K.K., Andersen O.J., Pogorelov T.V., Tajkhorshid E., Schiøtt B. Conformational dynamics of the human islet amyloid polypeptide in a membrane environment: toward the aggregation prone form. Biochemistry. 2016;55(13):2031–2042. doi: 10.1021/acs.biochem.5b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breydo L., Wu J.W., Uversky V.N. α-Synuclein misfolding and Parkinson's disease. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2012;1822(2):261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Jo E., McLaurin J., Yip C.M., George-Hyslop P.S., Fraser P.E. α-Synuclein membrane interactions and lipid specificity. J. Biol. Chem. 2000;275(44):34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 48.Meade R.M., Williams R.J., Mason J.M. A series of helical α-synuclein fibril polymorphs are populated in the presence of lipid vesicles. NPJ Parkinson's disease. 2020;6(1):1–5. doi: 10.1038/s41531-020-00122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fatafta H., Kav B., Bundschuh B.F., Loschwitz J., Strodel B. Disorder-to-order transition of the amyloid-β peptide upon lipid binding. Biophys. Chem. 2022;280 doi: 10.1016/j.bpc.2021.106700. [DOI] [PubMed] [Google Scholar]
- 50.Brender J.R., Salamekh S., Ramamoorthy A. Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective. Acc. Chem. Res. 2012;45(3):454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burré J., Sharma M., Südhof T.C. α-Synuclein assembles into higher-order multimers upon membrane binding to promote SNARE complex formation. Proc. Natl. Acad. Sci. U. S. A. 2014;111(40):E4274–E4283. doi: 10.1073/pnas.1416598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer A., Roeters S.J., Schilderink N., Hommersom B., Heeren R.M., Woutersen S., Claessens M.M., Subramaniam V. The impact of N-terminal acetylation of α-synuclein on phospholipid membrane binding and fibril structure. J. Biol. Chem. 2016;291(40):21110–21122. doi: 10.1074/jbc.M116.726612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fusco G., De Simone A., Gopinath T., Vostrikov V., Vendruscolo M., Dobson C.M., Veglia G. Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat. Commun. 2014;5(1):1–8. doi: 10.1038/ncomms4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sellin D., Yan L.-M., Kapurniotu A., Winter R. Suppression of IAPP fibrillation at anionic lipid membranes via IAPP-derived amyloid inhibitors and insulin. Biophys. Chem. 2010;150(1-3):73–79. doi: 10.1016/j.bpc.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Cawood E.E., Karamanos T.K., Wilson A.J., Radford S.E. Visualizing and trapping transient oligomers in amyloid assembly pathways. Biophys. Chem. 2021;268 doi: 10.1016/j.bpc.2020.106505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Najarzadeh Z., Pedersen J.N., Christiansen A.B., Shojaosadati S.A., Pedersen J.S., Otzen D.E. Bacterial amphiphiles as amyloid inducers: effect of rhamnolipid and lipopolysaccharide on FapC fibrillation. Biochim. Biophys. Acta Proteins Proteom. 2019;1867 doi: 10.1016/j.bbapap.2019.140263. [DOI] [PubMed] [Google Scholar]
- 57.Bleem A., Christiansen G., Madsen D.J., Maric H., Strømgaard K., Bryers J.D., Daggett V., Meyer R.L., Otzen D.E. Protein engineering reveals mechanisms of functional amyloid formation in Pseudomonas aeruginosa biofilms. J. Mol. Biol. 2018;430(20):3751–3763. doi: 10.1016/j.jmb.2018.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Najarzadeh Z., Mohammad-Beigi H., Nedergaard Pedersen J., Christiansen G., Sønderby T.V., Shojaosadati S.A., Morshedi D., Stromgaard K., Meisl G., Sutherland D., Skov Pedersen J., Otzen D.E. Plant polyphenols inhibit functional amyloid and biofilm formation in pseudomonas strains by directing monomers to off-pathway oligomers. Biomolecules. 2019;9(11):659. doi: 10.3390/biom9110659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders H.M., Jovcevski B., Marty M.T., Pukala T.L. Structural and mechanistic insights into amyloid-β and α-synuclein fibril formation and polyphenol inhibitor efficacy in phospholipid bilayers. FEBS J. 2022;289(1):215–230. doi: 10.1111/febs.16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engel M.F., VandenAkker C.C., Schleeger M., Velikov K.P., Koenderink G.H., Bonn M. The polyphenol EGCG inhibits amyloid formation less efficiently at phospholipid interfaces than in bulk solution. J. Am. Chem. Soc. 2012;134(36):14781–14788. doi: 10.1021/ja3031664. [DOI] [PubMed] [Google Scholar]
- 61.Beasley M., Stonebraker A.R., Hasan I., Kapp K.L., Liang B.J., Agarwal G., Groover S., Sedighi F., Legleiter J. Lipid membranes influence the ability of small molecules to inhibit huntingtin fibrillization. Biochemistry. 2019;58(43):4361–4373. doi: 10.1021/acs.biochem.9b00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.