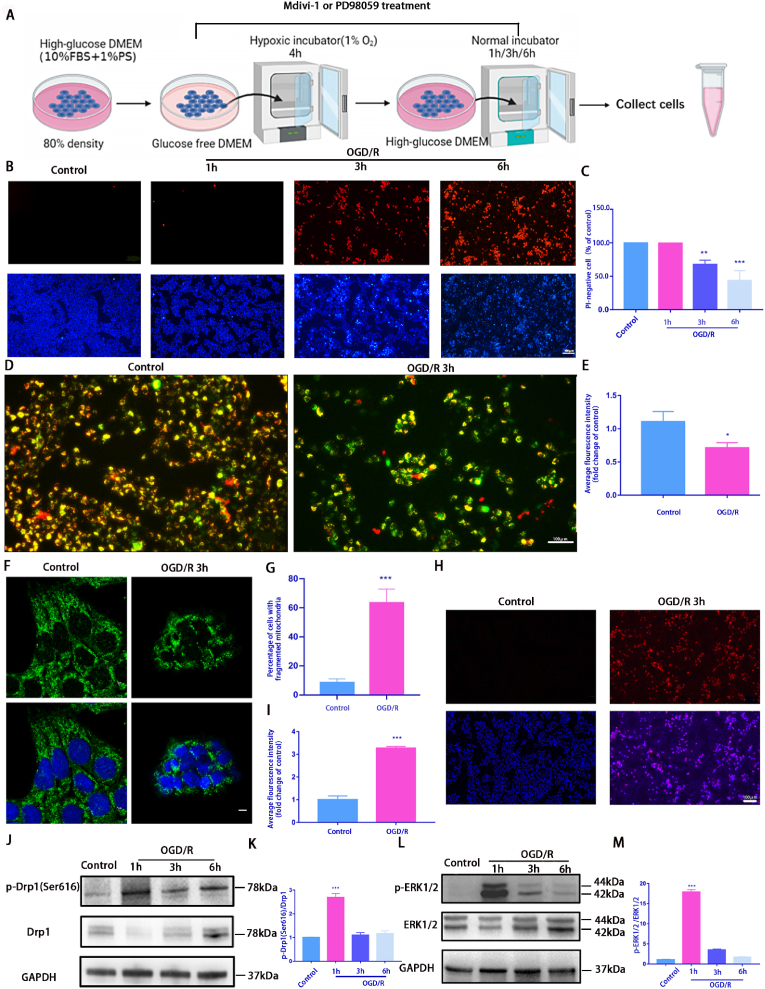
Fig. 1.
OGD/R model induces mitochondrial damage and death in R28 cells
A: A schematic diagram of OGD/R treatment. R28 cells cultured in glucose-free DMEM were incubated in a hypoxic chamber (1% O2) for 4 h followed by 1 h, 3 h, or 6 h reoxygenation in normal complete medium. Control cells were maintained in a complete medium under normoxic conditions. Cells from each time point were used for subsequent analysis; B: OGD/R-induced cell death was detected using Hoechst 33342 and PI dual staining. Scale bar = 100 μm; C: Quantifying the percentage of PI-positive cells(n = 9); D: The ΔψM was measured using JC-1 staining(2.0 μg/mL) after 3h OGD/R. Scale bar = 100 μm; E: The relative red/green ratio quantification(n = 9); F: Confocal microscopy was used to determine mitochondrial morphology using Tom20 antibodies via immunofluorescence after 3 h OGD/R. Scale = 2 μm; G: The average mitochondrial length was analyzed(n = 9); H: Mito-SOX(2 μM) staining was used to detect the ROS level of mitochondrial after 3h OGD/R. Red fluorescence represents mitochondrial ROS production, and Hoechst staining in the nuclei is shown in blue. Scale bar = 100 μm; I: Relative Mito-SOX red fluorescence quantification(n = 9); J and L: Western blotting was performed to detect p-Drp1(Ser616), Drp1, p-ERK1/2, and ERK1/2 expression at each time point after OGD/R treatment; K and M: Quantification of p-Drp1(Ser616), Drp1, p-ERK1/2, and ERK1/2 levels. Relative protein levels were calculated using ImageJ software(n = 9). Values are expressed as means ± standard deviations (SD). One-way analysis of variance (ANOVA) was followed by the Dunnett's test. *P < 0.05, **P < 0.01, ***P < 0.001and****P < 0.0001 vs Control.