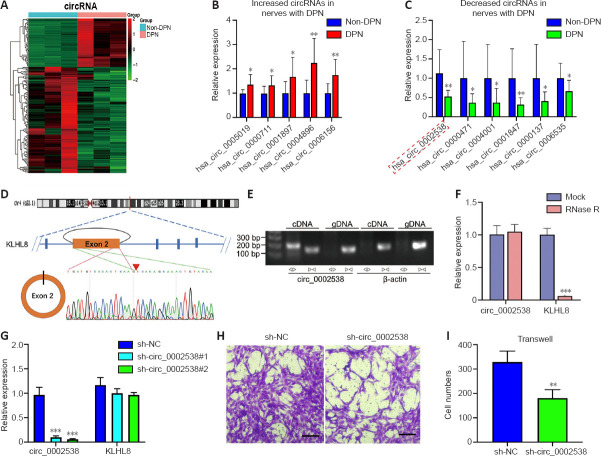
Figure 3.
Characterization of circ_0002538 and its function in SCs.
(A) Hierarchical clustering analyses of DEcircRNAs (n = 3). (B, C) RT-PCR verified five circRNAs with upregulated expression and six circRNAs with downregulated expression, and the results were consistent with the RNA-seq data (n = 12). The red dotted box highlights the circRNA of interest. Y-axis: Fold changes in circRNA expression compared with the non-diabetic group. *P < 0.05, **P < 0.01, vs. non-diabetic group (independent-sample t-test). (D) Schematic diagram showing that circ_0002538 was formed by the circularization of KLHL8 exon 2. The red arrow represents the “head-to-tail” splicing site of circ_0002538, confirmed by Sanger sequencing. (E) We used divergent primers and convergent primers to amplify circ_0002538 in cDNA and gDNA. We used β-actin as a negative control. (F) circ_0002538 and KLHL8 mRNA in SCs were detected via RT-PCR after incubation with or without RNase R. Y-axis: fold changes in RNA expression compared with the mock group. ***P < 0.001, vs. mock group (independent-sample t-test). (G) circ_0002538 and KLHL8 mRNA levels were evaluated in the sh-circ_0002538-transfected SCs via RT-PCR. Y-axis: fold changes in RNA expression compared with the sh-NC group. ***P < 0.001, vs. sh-NC group. (H, I) The migrating number of SCs in the sh-circ_0002538 group was lower than that in the sh-NC group in the Transwell assays. **P < 0.01, vs. sh-NC group (independent-sample t-test). Scale bars: 100 μm. All bar graphs represent the average of at least three independent replicates, and the error bars are the SD. cDNA: Complementary DNA; DPN: diabetic peripheral neuropathy; gDNA: genomic DNA; KLHL8: Kelch-like family member 8; sh-circ_0002538: short hairpin RNA for circ_0002538; sh-NC: normal control for short hairpin RNA.