Figure 7.
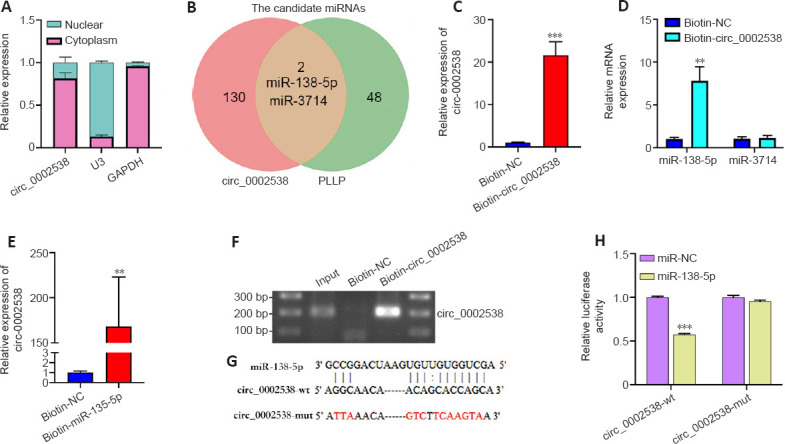
circ_0002538 acts as a sponge for miR-138-5p in SCs.
(A) Nuclear and cytoplasmic separation assays detecting the localization of circ_0002538 in SCs. Y-axis: proportion of nuclear and cytoplasmic RNA to total RNA. (B) Venn diagram showing the overlap of circ_0002538 candidate miRNAs and PLLP candidate miRNAs. (C) circ_0002538 was pulled down in SC lysates by the biotin-circ_0002538 probe and detected via RT-PCR. The relative level of circ_0002538 was normalized to the input. Y-axis: fold changes in circ_0002538 expression compared with the biotin-NC group. ***P < 0.001, vs. Biotin-NC group (independent-sample t-test). (D) miR-138-5p was pulled down by the biotin-circ_0002538 probe, while miR-3714 was not, as shown by the RT-PCR. Y-axis: fold changes in miRNAs expression compared with the biotin-NC group. **P < 0.01, vs. Biotin-NC group (independent-sample t-test). (E, F) circ_0002538 was pulled down in SC lysates by the biotin-miR-138-5p probe, as shown by RT-PCR. The relative level of circ_0002538 was normalized to the input. Y-axis: fold changes in circ_0002538 expression compared with the biotin-NC group. **P < 0.01, vs. Biotin-NC group (independent-sample t-test). (G) The miR-138-5p binding site of circ_0002538 was predicted via RNAhybrid. The mutant sequences are marked in red. (H) Dual-luciferase reporter assays of HEK293T cells cotransfected with miR-138-5p mimics, circ_0002538 wild-type (circ_0002538-wt), or circ_0002538 mutant type (circ_0002538-mut) plasmids. Y-axis: relative luciferase activity compared with the miR-NC + circ_0002538-wt group. ***P < 0.001, vs. miR-NC group (one-way analysis of variance and Tukey’s post hoc test). All bar graphs represent the average of three independent replicates, and the error bars are the SD. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; mut: mutant type; NC: normal control; PLLP: plasmolipin; U3: small nucleolar U3 RNA; wt: wild-type.
