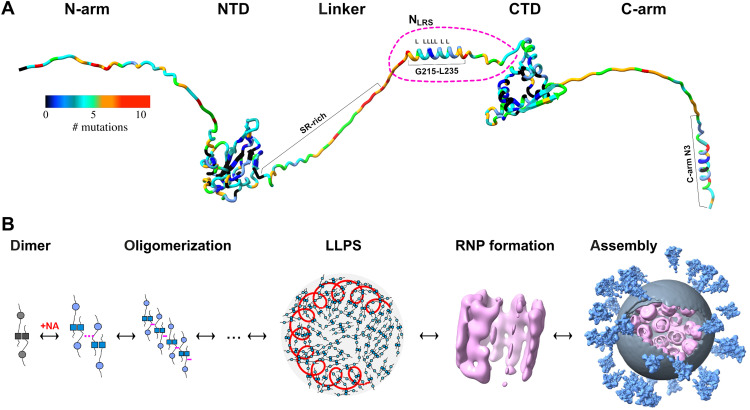
Fig. 1. Schematics of N-protein structure and assembly.
(A) N-protein with folded domains (NTD and CTD) and IDRs (N-arm, linker, and C-arm; all IDRs are artificially stretched for clarity). The variability of the amino acid sequence is highlighted through colors indicating for each position the number of distinct mutations contained in the GISAID genomic data base (as of July 2022) (21, 55, 72). (B) Current model for viral assembly: N-protein dimers, noncovalently linked in the CTD (dark squares), undergo a conformational change upon NA binding that allows oligomerization and promotes LLPS through weak protein-protein and/or protein-NA interactions. The dense phase containing genomic RNA (red coil) permits formation of RNPs [Electron Microscopy Data Bank (EMDB) 11868 (16)] leading to viral assembly [EMDB 30430 (17)].