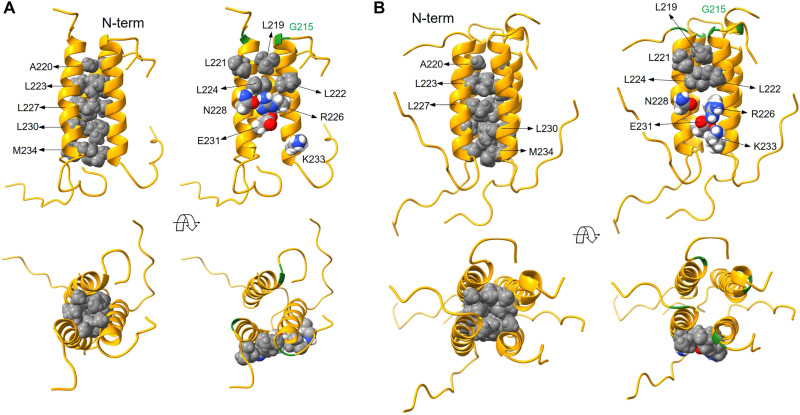
Fig. 6. MD simulations highlight residues that stabilize NLRS oligomers.
Shown are snapshots from the MD simulations for the trimer (A) and tetramer (B). The cores of the oligomers are formed by five nonpolar residues in a well-packed configuration (left column of each panel). More specific interactions stabilizing the complex surfaces are (right column of each panel): a hydrophobic surface cluster formed by four leucine residues, two salt bridges (R226-E231 and K233-E231), and an H-bond interaction between polar (N228) and charged (R226) residues. Helix backbones drawn as gold ribbons; interacting residues rendered as atom-based Van der Waals spheres (nonpolar residues in gray; polar and charged residues in color: white, H; red, O; gray, C; blue, N). G215 is shown in green for reference.