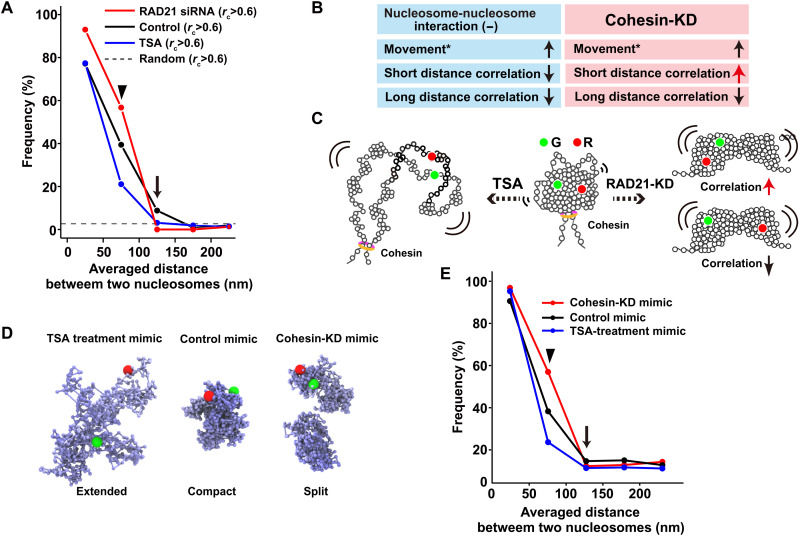
Fig. 6. Distinct roles of nucleosome-nucleosome interactions and cohesin during condensed domain formation.
(A) Frequency plots of the nucleosome pairs with rc > 0.6 (congruence coefficient rc) versus the pair distances in living siRAD21-treated (cohesin-KD) HeLa cells (red), untreated cells (black), and TSA-treated cells (blue). The dashed line shows the frequency of randomly collected nucleosome pairs with rc > 0.6. Note that closely located nucleosomes (<50 nm) showed high correlations even after TSA treatment since this distance corresponds to a few nucleosomes, possibly with a connected chain. (B) Summary of results in (A). Movement data (*) were obtained from (18, 71). (C) Simplified schematic of possible TSA-treated (left) and cohesin-KD (right) domains. (D) Computational models that mimic TSA-treated, untreated control, and cohesin-KD domains (from left to right). For the cohesin-KD mimic, two split domains were created. (E) Frequency plots of the nucleosome pairs with rc > 0.2 (congruence coefficient rc) versus the averaged pair distances in the computationally modeled domains of TSA-treatment mimic (red), untreated control mimic (black), and cohesin-KD mimic (blue) shown in (D).