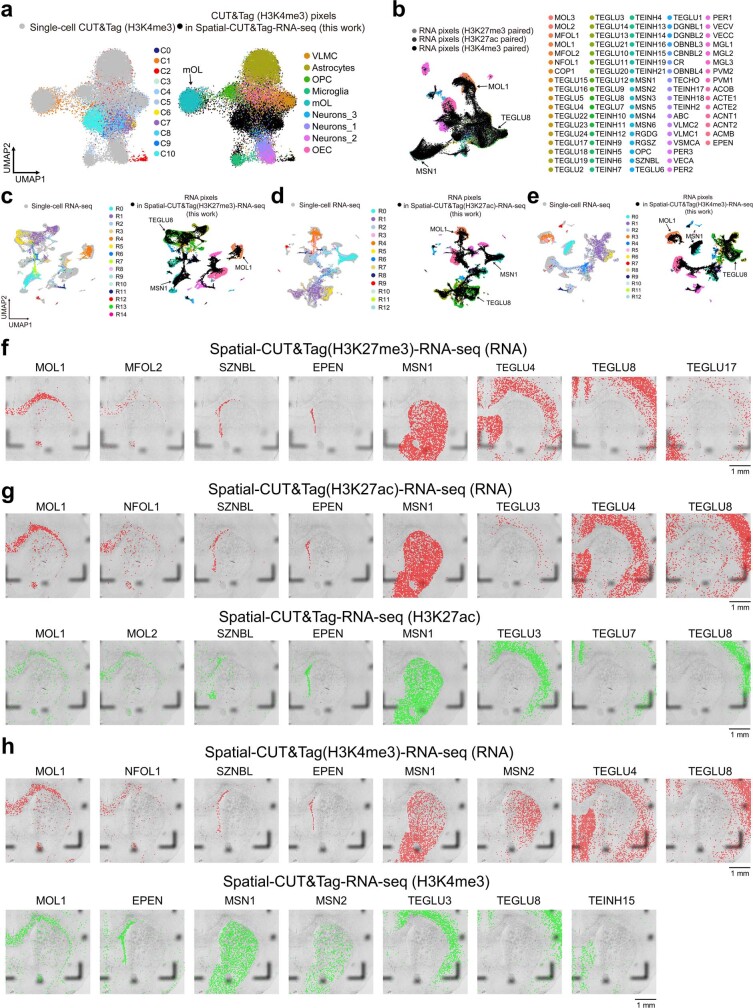
Extended Data Fig. 7. Further analysis for spatial-CUT&Tag-RNA-seq with P22 mouse brain.
a, Integration of CUT&Tag (H3K4me3) data in spatial-CUT&Tag-RNA-seq with scCUT&Tag (H3K4me3) data36 from mouse brain. b, Integration of RNA data in spatial- CUT&Tag(H3K27me3)-RNA-seq, spatial-CUT&Tag(H3K27ac)-RNA-seq, and spatial-CUT&Tag(H3K4me3)-RNA-seq with scRNA-seq data32 from mouse brain. c-e, Integration of RNA data in spatial-CUT&Tag(H3K27me3)-RNA-seq (c), RNA data in data in spatial-CUT&Tag(H3K27ac)-RNA-seq (d), and RNA data in spatial-CUT&Tag(H3K4me3)-RNA-seq (e) with scRNA-seq data32 from mouse brain. f, Spatial mapping of cell types identified by label transfer from scRNA-seq32 to RNA data in spatial-CUT&Tag(H3K27me3)-RNA-seq. g, Spatial mapping of cell types identified by label transfer from scRNA-seq32 to RNA (top) and from scRNA-seq32 to CUT&Tag (H3K27ac, bottom) data in spatial-CUT&Tag(H3K27ac)-RNA-seq. h, Spatial mapping of cell types identified by label transfer from scRNA-seq32 to RNA (top) and from scRNA-seq32 to CUT&Tag (H3K4me3, bottom) data in spatial-CUT&Tag(H3K4me3)-RNA-seq.