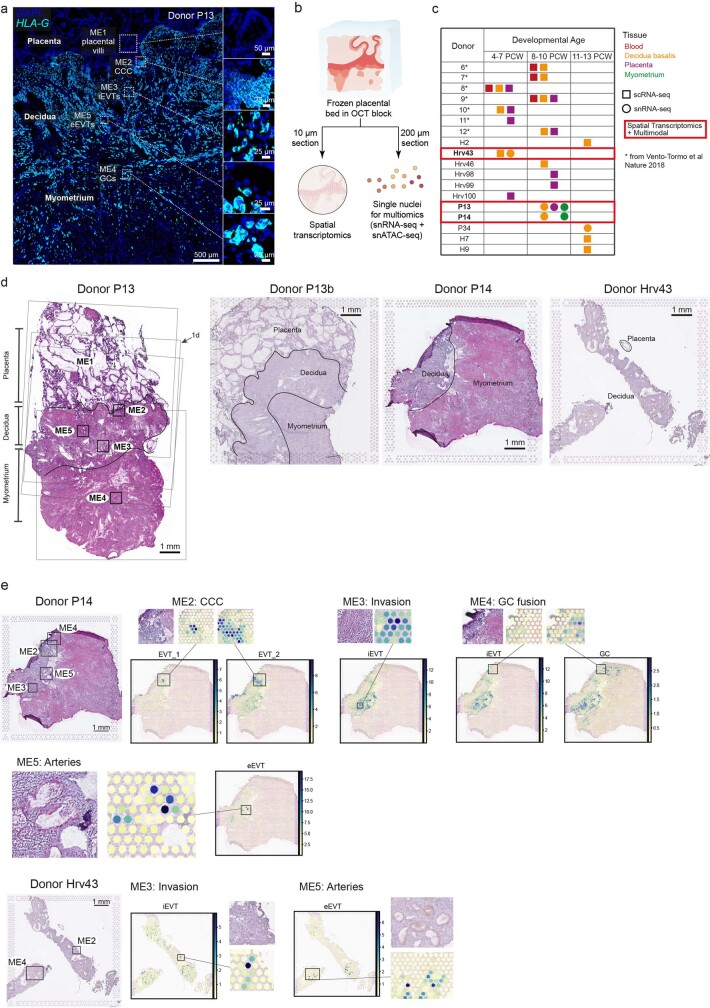
Extended Data Fig. 1. Spatial transcriptomics of human placental bed.
a: High-resolution imaging of a section of the placenta-decidua interface stained by in situ hybridization (smFISH) for HLA-G, illustrating the depth of invasion of EVTs into the uterus (n = 1). Magnified insets (dashed squares) highlight the HLA-G-negative placental villi, and HLA-G+ EVTs emerging from the CCC to invade the decidua and myometrium. b: Overview of experimental design of the study. c: Cohort composition split by gestational age window (post-conceptional weeks, PCW) representing tissues sampled from each donor and performed assays. Highlighted in red rectangles are the three donors whose tissues have been additionally profiled with spatial transcriptomics (Visium) and multiome assays. d: Histological overview (H&E staining) of donors P13, P14 and Hrv43 tissues with annotations of tissue regions. For the implantation site of donor P13 (~ 8-9 PCW, left); black squares (small) indicate trophoblast microenvironments in space; faint grey squares (large) indicate positioning of tissue on Visium spatial transcriptomics capture areas; arrow indicates representative Visium section further explored in Fig. 1d. For Visium, P13 (n = 5 feature areas, 4 consecutive slides with overlapping positions and 1 slide from an additional tissue block - P13b), P14 (n = 2 feature areas, consecutive slides with same position), Hrv43 (n = 1 feature area). e: Cell state locations (derived with cell2location) in representative Visium sections of donors P14 and Hrv43 highlighting relevant spatial trophoblast microenvironments. Spot colour indicates cell state densities computed by cell2location as the number of cells of a given cell state in a Visium spot. Cytotrophoblast cell column (CCC), extravillous trophoblast (EVT), interstitial EVT (iEVT), giant cells (GC), endovascular EVT (eEVT), single-cell RNA sequencing (scRNA-seq), single-nuclei RNA sequencing (snRNA-seq), microenvironment (ME), Hematoxylin and Eosin (H&E).