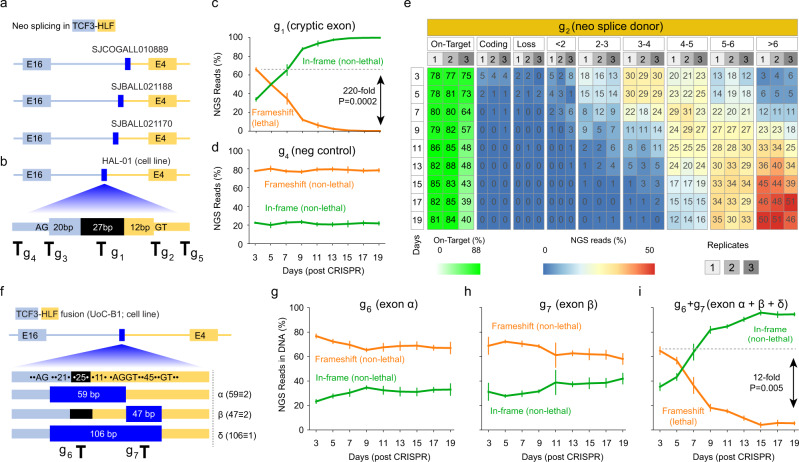
Fig. 6. Neo-splicing in oncogenic fusions and genome-editing-based therapeutic targeting.
a In our cohort, all samples with TCF3-HLF fusions harbor neo-splicing events due to incompatible exon frames between TCF3 exon 16 and HLF exon 4 (Supplementary Fig. 7e). b We confirmed B-ALL cell line HAL-01 (DSMZ#: ACC 610) also harbors this pattern and designed guide RNAs to target the cryptic exon (g1) and the neo-splice sites (g2 and g3) as well as negative control guides (g4: 199 bps upstream of g3; g5: 52 bps downstream of g2). Black shading indicates non-template insertion sequence (27 bp). c Cryptic exon is essential to HAL-01 by CRIPSR targeting using guide g1, which leads to a 220-fold decrease of cells with lethal editing (two-sided t-test; P value = 0.0002; n = 3). Shown are percentages (y-axis) of putative lethal (orange) and non-lethal (green) editing measured using NGS reads as a function of time from day 3 to day 19 (x-axis) post editing for three replicates (error bars indicate standard deviation). Indels leading to frameshift of fusion transcripts are called lethal and in-frame indels are called non-lethal. d Negative control guide (g4) that targets the upstream intronic region of the cryptic exon. Similar as panel c, percentage of putative lethal (frameshift; orange) and non-lethal (in-frame; green) editing measured by using NGS reads are shown from day 3 to day 19. e Neo-splice donor is essential to HAL-01 by CRISPR targeting using guide g2. The induced indels that happened to fall into the coding region and lead to frameshift of TCF3-HLF are categorized into “Coding” group. Indels that directly disrupt the splice donor site are categorized into “Loss” group. Most of the induced indels leave a residual GT that may still serve as a splice donor. The binding affinity of these residual donors is calculated using the position weight matrix (PWM) approach (see Methods) and the binding affinity scores are categorized into different bins (<2, 2–3, etc.). The percentage of NGS reads carrying induced indels are calculated for each bin from day 3 to day 19 post editing for three replicates. Also illustrated are on-target editing rate (green heatmap). f CRISPR targeting in the presence of alternative splicing. B-ALL cell line UoC-B1 also harbors TCF3-HLF fusion. However, in this fusion, we detected three neo-splicing patterns, α, β, and δ, where the first two splicing patterns can generate in-frame fusion proteins in the parental cells and δ cannot. We designed two guides (g6 and g7) to test the potential compensatory function of these isoforms. g, h Single guide editing led to marginal depletion of edited cells from day 3 to day 19 for putative lethal indels (orange) that can disrupt corresponding transcripts. i Double guide editing. Indels with putative lethal effect (orange) demonstrated a quick decrease (12-fold; two-sided t-test; P value = 0.005; n = 3) in abundance from day 3 to day 19, while indels with putative non-lethal effect demonstrated an increasing abundance. Data value and error bar at each time point represent mean of putative indels (orange for lethal; green for non-lethal as control) and standard deviation from three replicates in panels c and d and panels g–i. Source data are provided accordingly as sheets a, b and c–e in Source Data file.