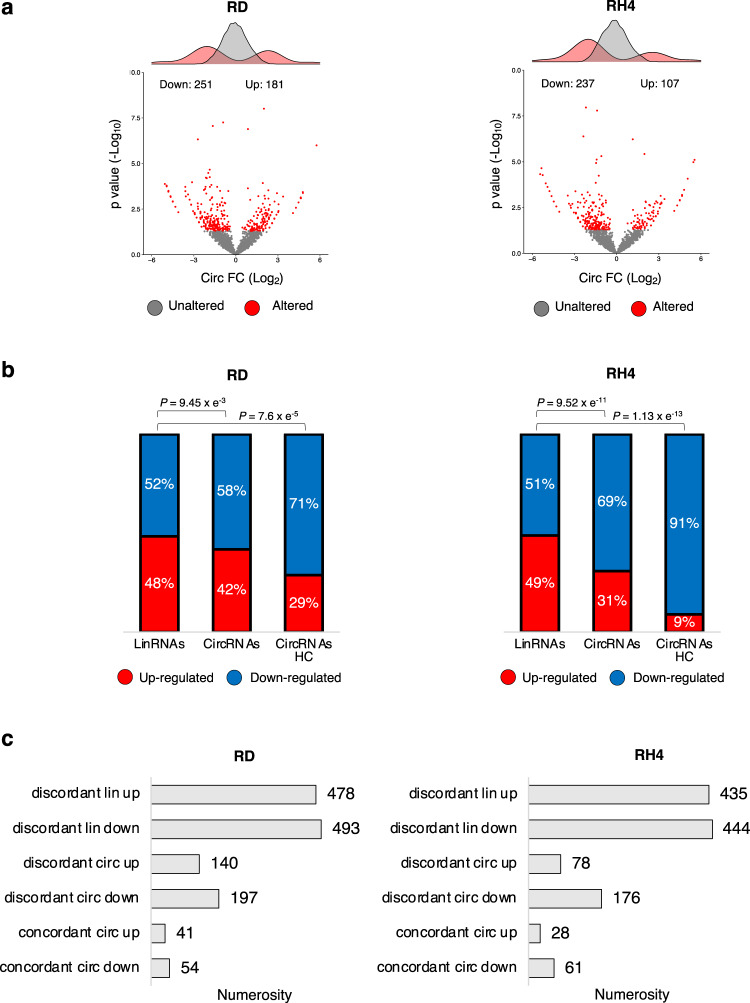
Fig. 3. YTHDC1 depletion downregulates circRNAs in RMS.
a Volcano plots showing for each circRNA identified in the RNA-seq experiment the log2 fold change and the −log10 p value upon YTHDC1 knock-down in RD (left panel) or in RH4 (right panel) cells. The distributions of the fold change values are shown above the volcano plots. Significantly altered circRNAs (p value < 0.05) or unaltered circRNAs are indicated by red or gray dots, respectively. See the methods section for statistical analyses details. b Stacked bar charts with percentage of down- and upregulated linear (“LinRNAs”), circRNAs (“CircRNAs”), or “high-confidence” circRNAs (“CircRNAs HC”) upon YTHDC1 knock-down in RD (left panel) or RH4 (right panel) cell lines. P values for the differences between proportions were calculated using Fisher exact two-tailed test. c Bar-plots depicting the numerosity of different scenarios of deregulation in RD (left panel) or in RH4 (right panel) cells upon YTHDC1 knock-down. Source data are provided as a Source Data file.