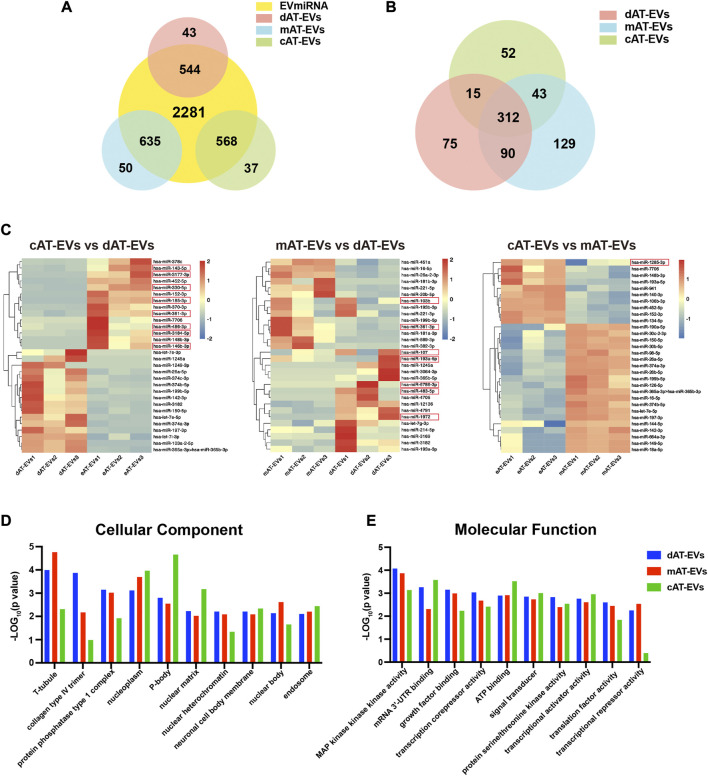
FIGURE 9.
Bioinformatics analysis of miRNA enriched in AT-EVs (A) Venn diagram illustrating the number of miRNAs in dAT-EVs, mAT-EVs, and cAT-EVs, respectively, overlapping with those in the EVmiRNA database. (B) Venn diagram illustrating the number of overlapping miRNAs between dAT-EVs, mAT-EVs, and cAT-EVs (C) Hierarchical clustering of differentially expressed miRNAs (fold change >2.0 and p < 0.01; unreported miRNAs, as well as those with low TPM values, were omitted) between cAT-EVs and dAT-EVs (left), mAT-EVs and dAT-EVs (middle), and cAT-EVs and mAT-EVs (right), respectively. Rows represent miRNAs, whereas columns represent individual replicates. (D–E) Gene ontology (GO) analysis of predicted differentially expressed genes of cellular components and molecular functions, based on the identified differentially expressed miRNAs.