Abstract
Rationale and Objectives:
Accurate segmentation of the upper airway lumen and surrounding soft tissue anatomy, especially tongue fat, using magnetic resonance images is crucial for evaluating the role of anatomic risk factors in the pathogenesis of obstructive sleep apnea (OSA). We present a convolutional neural network to automatically segment and quantify upper airway structures that are known OSA risk factors from unprocessed magnetic resonance images.
Materials and Methods:
Four datasets (n = [31, 35, 64, 76]) with T1-weighted scans and manually delineated labels of 10 regions of interest were used for model training and validations. We investigated a modified U-Net architecture that uses multiple convolution filter sizes to achieve multi-scale feature extraction. Validations included four-fold cross-validation and leave-study-out validations to measure generalization ability of the trained models. Automatic segmentations were also used to calculate the tongue fat ratio, a biomarker of OSA. Dice coefficient, Pearson’s correlation, agreement analyses, and expert-derived clinical parameters were used to evaluate segmentations and tongue fat ratio values.
Results:
Cross-validated mean Dice coefficient across all regions of interests and scans was 0.70 ± 0.10 with highest mean Dice coefficient in the tongue (0.89) and mandible (0.81). The accuracy was consistent across all four folds. Also, leave-study-out validations obtained comparable accuracy across uniquely acquired datasets. Segmented volumes and the derived tongue fat ratio values showed high correlation with manual measurements, with differences that were not statistically significant (p < 0.05).
Conclusion:
High accuracy of automated segmentations indicate translational potential of the proposed method to replace time consuming manual segmentation tasks in clinical settings and large-scale research studies.
Keywords: Upper airway, obstructive sleep apnea, deep learning, segmentation, tongue fat ratio
INTRODUCTION
Obstructive sleep apnea (OSA) is a common sleep disorder that is characterized by recurrent episodes of partial or complete collapse of the upper airway during sleep. (1,2) Obesity is the primary risk factor for OSA due to the increased volume of the surrounding soft tissues of the upper airway. In particular, Schwab et al showed that increased volume of the lateral pharyngeal walls, total soft tissue, and tongue could compromise upper airway caliber and thereby increase the risk of developing OSA. (3) Moreover, studies have also shown that tongue fat is increased in patients with sleep apnea and that reductions in tongue fat volume are a primary mediator of improvements in OSA severity seen with weight loss. (4,5) As a result, quantification of the volumes of these clinically relevant upper airway structures is critical for comprehensive investigation of the pathogenesis of OSA.
Magnetic resonance imaging (MRI) has traditionally been used to quantify the volume of these upper airway anatomy, including airway sizes, craniofacial traits, and soft tissues. (3) Success of imaging studies largely depends on segmentation accuracy. The current gold standard for segmentation is manual delineation of upper airway structures on T1-weighted MR scans. This is a time-consuming task, which is not feasible for large-scale datasets such as electronic health record-linked or radiologic biorepositories. Additionally, as MRI scanners produce 3D images by generating multiple 2D slices, radiologists or medical experts are required to go through the 3D dataset slice-by-slice, which may lead to inconsistencies in the 3D contours of manually segmented structures. When relying on manual scoring, intra and inter-rater variability can be high due to factors such as lack of contrast in boundaries that separate structures, small size of target structures, high anatomical variations of upper airway structures across subjects, and scanner related variations. To minimize this variability among and within raters, as well as to protect against drifts in scoring over time, specialized training and ongoing data quality assessments are required.
Deep learning (DL) has been on the cutting-edge of machine learning research in recent years, consistently and significantly outperforming traditional methods in a variety of segmentation tasks. (6) Recent advances in DL methods, specifically using convolutional deep neural networks (CNNs), have led to a major paradigm shift in image classification and segmentation. Traditional methods often rely on extensive and hand-crafted pre-processing, feature engineering, and reduction techniques to transform initial 2D or 3D image data into a set of optimal features that are then presented as training input to a subsequent classifier. In contrast, DL methods can directly operate on raw or minimally pre-processed data and learn complex multi-variate patterns in the data through iterative training and optimization of network weights in multiple hidden layers. CNNs are specifically designed to analyze 2D or 3D images. An input image passes through a series of convolution layers followed by pooling layers, which act together as filters to extract a very large number of translation invariant local features in different scales, without need for the manual feature engineering traditionally required. The network learns the optimal weights (i. e., the combination of optimal filters) that minimize the loss function, through intensive training with back-propagation. For example, CNN architectures have obtained state-of-the-art performance in various neuroimaging segmentation tasks, such as brain extraction and segmentation of abnormal tissues (white matter lesions, infarcts, brain tumors). (7,8) As such, these cutting-edge approaches are ideally suited to automate upper airway MRI analysis.
To the best of our knowledge, there are no existing machine learning approaches that segment all the OSA-implicated upper airway structures identified by Schwab et al (3) There have only been methods that segment individual regions identified in Schwab’s study (3) in a semi-automatic or automatic manner. (9–14) Among these partial methods, all but three have been tested on cone-beam computed tomography. (12,13,14) However, cone-beam computed tomography results in low resolution scans and patient radiation, significantly limiting routine use in large studies. In recent years, MRI, which is a non-radiation based scanning method, has been used more frequently as it provides excellent contrast between upper airway structures.
Considering this development, Xie et al recently developed a semi-automatic anatomy-guided CNN methodology to segment the upper airway lumen on T2-weighted MR scans. (14) They split the upper airway into a subregion containing the nasal cavity and nasopharyngeal airway and another subregion containing the oropharyngeal, hypopharyngeal, and supraglottic/glottic laryngeal airway using a manual parcellation strategy. This strategy requires an expert to provide the initial location of the inferior, separation, and superior boundary slices of the upper airway. After automatically cropping the image based on dataset-specific spatial context as well as a normalization procedure, they train two 2D CNNs to segment each subregion and combine the two output masks into a final segmentation. While this work demonstrates a complete segmentation of upper airway lumen, it has three limiting factors in its applicability to OSA studies. First, this approach does not segment any of the important soft tissue structures surrounding the upper airway. Second, their use of a manual parcellation procedure necessitates human intervention for each scan, limiting its practicality when examining with large datasets. Third, their method only uses data from one site and an automatic cropping method trained on this specific dataset. This may prevent generalization due to differences in data acquisition from different locations.
A method that can automatically segment both upper airway lumen and the volume of the surrounding soft tissues implicated in OSA across different acquisition methods is necessary for large-scale studies investigating OSA pathogenesis; however, such methods are currently lacking. The present study describes a CNN-based DL method designed for automatically segmenting upper airway anatomic risk factors for OSA on T1-weighted MRI scans. We hypothesize that DL models that train on data with ground truth labels can accurately delineate upper airway structures, which can then be used to calculate volumetric features and derived imaging biomarkers that are not significantly different compared to those obtained from manual segmentations.
MATERIALS AND METHODS
Data Sources
The MRI datasets were provided by the researchers at the Division of Sleep Medicine at the University of Pennsylvania. The data included scans of 234 participants from four studies investigating phenotypic characteristics of sleep apnea patients (Study one: n = 43; Study two: n = 43; Study three: n = 72; Study four: n = 76), with samples comprising participants with and without OSA (based on sleep study results). The demographic breakdown of each study by gender, age, race/ethnicity, and OSA status is as follows: Study one (Gender: 45% Male/55% Female; Age (range): 48 (41–70); Race/ethnicity: 100% White; has OSA: 40%); Study two (Gender: 54% Male/46% Female; Age (range): 45 (28–72); Race/ethnicity: 44% African-American/47% White/9% Asian; has OSA: 55%); Study three (Gender: 100% Female; Age (range): 51 (45–54); Race/ethnicity: 45% African-American /48% White/7% Asian; has OSA: 80%); Study four (Gender: 62% Male/38% Female; Age (range): 41 (27–58); Race/ethnicity: 56% African-American/44% White; has OSA: 53%).
Subjects in studies one, two, and four were scanned by a Siemens Avanto 1.5T with 32-channel head/neck array while subjects in study three were scanned by a Siemens Prisma 1.5T with 32-channel head/neck array. For each subject, we had T1-weighted axial head and neck MRI with an image size of 512-by-512-by-58 voxels, a resolution of 0.508-by-0.508 mm, and 3 mm slice thickness. Manual delineation of each upper airway structure was performed by a trained expert using 2D axial slices, resulting in one 3D mask with multiple labels for each scan. There were 10 regions of interest (ROI): mandible, retropalatal airway, retroglossal airway, soft palate, tongue (genioglossus plus intrinsic muscles), “other tongue” (all other extrinsic tongue muscles, including digastric, mylohyoid, geniohyoid, styloglossus, and hyoglossus), epiglottis, parapharyngeal fat pads, retropalatal lateral walls, and retroglossal lateral walls. Figure 1 illustrates some target ROIs overlaid and labeled on a T1-weighted scan.
Figure 1.
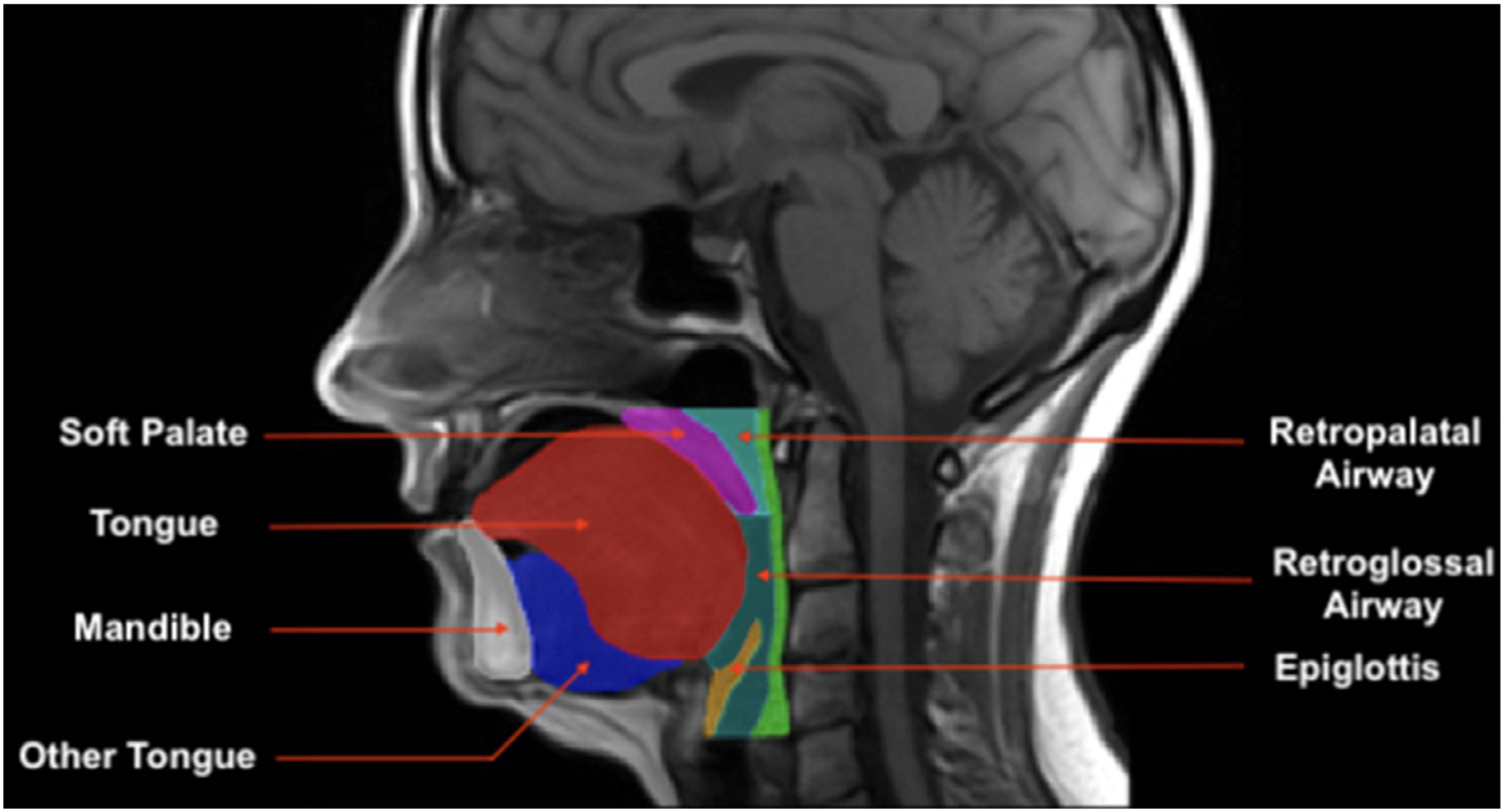
Target upper airway structures. Illustration of seven target upper airway structures overlaid on a single T1-weighted scan (midsagittal view).
MRI Quality Control and Pre-Processing
To create high quality testing and training sets, T1-weighted scans were manually verified for quality and 28 (~12%) scans of the initial dataset were excluded due to image processing errors (significant motion blur or artifacts), resulting in a final sample of 206 scans from independent subjects (Study one: n = 31; Study two: n = 35; Study three: n = 64; Study four: n = 76). Both in model training and testing phases, images were inputted to the network without any additional pre-processing steps. The main motivation for this design choice was to increase future applicability of the method in clinical settings by allowing the direct use of raw images from the scanner.
Network Model
We used a 2D convolution neural network that follows a modified U-Net architecture designed for generic application to neuroimaging segmentation tasks. (15) This model combines recently proposed advances in the field. The network architecture consists of an encoding path and a corresponding decoding path as in U-Net, but using ResNet and modified Inception-ResNet-A blocks in the encoding and decoding paths. (16,17,18) A voxel-wise, multi-class soft-max classifier layer produces class probabilities for each voxel independently. The architecture of the proposed model is shown in Figure 2.
Figure 2.
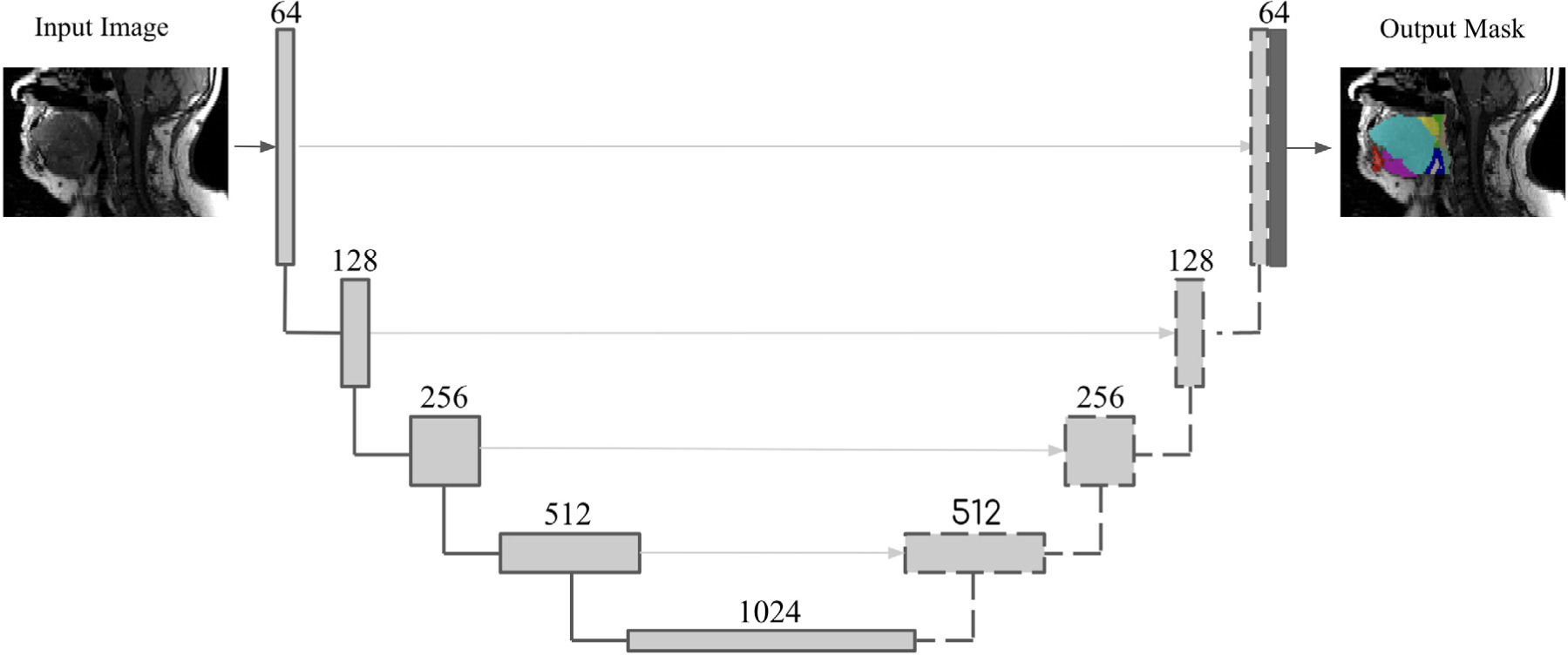
Network architecture to segment upper airway structures. The architecture of the convolutional neural network used for segmenting upper airway structures. Each item in the legend describes a unique and prototypical series of mathematical operations native to machine learning research. (13,14,15) Conv = convolutional layer; ReLU = rectified linear unit.
Training was performed on an NVIDIA TITAN Xp GPU with 12 GB memory. A learning rate of 0.05 was used with a decay of 0.98. A maximum epoch threshold was set at 200 epochs. Axial slices of 3D scans and their labels were used as independent samples during training, as well as during testing. On average, 45 epochs were used with batch size of three. The model was constructed in TensorFlow with the Adam optimizer (epsilon of 0.1) and a cross-entropy loss function. The average total training time was 25 hours for each model.
Tongue Fat Volume Ratio
Previous studies reported significant associations between increased tongue fat and OSA and suggested “tongue fat ratio” as an imaging biomarker for OSA. (4,19) Identification of decreasing muscle and increasing fat in the tongue has also been proposed as a predictor for presbyphagia. (19) We used the segmented tongue and fat pad ROIs to automatically estimate the tongue fat volume ratio of each subject. Since the intensity profile of tongue fat does not have a boundary that clearly delineates it from non-fat tongue regions, we measured tongue volume fat ratios by normalizing the intensity values in the segmented tongue by the average intensity in the segmented fat pads region, which is used as a reference for the expected intensity of fat tissues. Normalized values within the tongue were thresholded to exclude non-fat regions and were integrated to estimate the final tongue fat volume ratio. We compared the estimated tongue fat volume ratios to those calculated from manual segmentations of the tongue and the fat pads. In our cross-validation experiments, we investigated the correlation and statistical difference between the two adjusted tongue fat volume ratios (voxel-weighted at two different thresholds: max [“V1”] and 99th percentile fat pad intensities [“V2”]) calculated from our model-derived segmentations to those calculated from manual segmentations.
Evaluation and Metrics
The model was applied in two distinct experiments. First, we performed a four-fold cross validation, with 25% of the data left out for testing (n ≈ 51) and the remaining 75% used for training (n ≈ 155) for each fold. Then, we also performed “leave-study-out” validations. We created four models, each one trained with three studies and one unique study left out for testing. For quantitative evaluations in both experiments, output multi-class segmentation maps from each fold were pooled together and labels for each structure were compared to reference manual segmentations — considered as the gold standard — using standard metrics for comparison of binary segmentations. As two complementary metrics for accuracy evaluation, we reported the mean and one standard deviation of Dice scores across all subjects (described in more details below).
Spatial Overlap-Based Metric (Dice Score)
The primary metric of accuracy of segmentation labels was the Dice score, which was calculated individually for each different label class. In the context of biomedical imaging, the Dice score is used as a statistical validation metric to quantify the amount of spatial overlap between two segmentation masks. Given two sets of voxels S1 and S2 (i.e., two segmentation masks), the Dice score is defined as:
Correlational Analysis
Dice metrics are very sensitive to overall overlap between ground-truth and automated segmentations. However, in most analyses, the volume of the segmented region is the primary outcome of interest. Accordingly, we calculated, independently for each structure, the correlation between the ROI volumes obtained from ground-truth and automated segmentations across subjects.
Agreement Analysis
To understand the agreement between measures derived from the ground-truth and DL methods, we conducted analyses described by Bland and Altman, (20) including evaluations of the mean difference between techniques (bias), calculation of limits of agreement (equal to ± 2 standard deviations around the mean difference), and evaluation of the correlation between the average value of the two methods and the mean difference. We defined a priori the limits of maximum acceptable differences based on segmentation training standards – defined as ± 5% of 2 standard deviations above the mean of the gold standard segmentations’ volumes (Table 1). We compared the limits of agreement with the a priori defined limits to determine if the results were within clinically acceptable parameters.
TABLE 1.
The Expert-Defined Limits of Acceptable Differences for Each of the 10 Regions of Interest Under Consideration
| Limits of Maximum Acceptable Differences for each Region of Interest | |
|---|---|
| Structure | Maximum Acceptable Differences (in mm3) |
| Mandible | ±4050 |
| Retropalatal airway | ±358 |
| Retroglossal airway | ±659 |
| Soft palate | ±705 |
| Tongue | ±6041 |
| Other tongue | ±2547 |
| Epiglottis | ±159 |
| Fat pads | ±400 |
| Retropalatal lateral walls | ±1050 |
| Retroglossal lateral walls | ±870 |
These limits are defined as ±5% of two standard deviations above the mean of the 206 manual segmentations volumes.
RESULTS
Study Cohorts
Images utilized in this analysis came from four different cohorts of patients, as described in Data Sources. Mean ages across the cohort ranged from 41–51 years-old. Overall, 38% of the cohort was male, 39% were African American, 57% were White, and 4% Asian, and 59% had OSA. Study 1 included only White participants and Study three included only females.
Accuracy of the Deep Learning Algorithm
We evaluated the accuracy of the DL generated segmentations compared to gold-standard manual segmentations using Dice scores, as detailed in the Methods. In cross-validated segmentation experiments, our model obtained an overall spatial overlap accuracy of 0.70 ± 0.10 (Table 2). Fold-aggregated mean Dice scores for each ROI are visualized via boxplot (Fig 3).
TABLE 2.
Mean Dice Scores for the Cross-Validated Experiment Reported Across All n = 206 Subjects (Along With One Standard Deviation)
| Mean (Standard Deviation) Dice Scores for the Cross-Validated Experiments | |||||
|---|---|---|---|---|---|
| Structure | Fold 1 (n = 51) | Fold 2 (n = 51) | Fold 3 (n = 52) | Fold 4 (n = 52) | Averaged |
| Mandible | 0.81 (0.06) | 0.81 (0.07) | 0.81 (0.07) | 0.82 (0.06) | 0.81 (0.07) |
| Retropalatal airway | 0.62 (0.12) | 0.64 (0.12) | 0.63 (0.13) | 0.64 (0.11) | 0.63 (0.12) |
| Retroglossal airway | 0.72 (0.10) | 0.73 (0.09) | 0.73 (0.09) | 0.74 (0.10) | 0.73 (0.10) |
| Soft palate | 0.69 (0.13) | 0.66 (0.14) | 0.67 (0.12) | 0.67 (0.12) | 0.67 (0.13) |
| Tongue | 0.89 (0.04) | 0.89 (0.04) | 0.89 (0.04) | 0.89 (0.04) | 0.89 (0.04) |
| Other tongue | 0.66 (0.12) | 0.67 (0.11) | 0.69 (0.10) | 0.68 (0.11) | 0.68 (0.11) |
| Epiglottis | 0.61 (0.10) | 0.59 (0.14) | 0.63 (0.14) | 0.57 (0.13) | 0.60 (0.13) |
| Fat pads | 0.57 (0.12) | 0.57 (0.13) | 0.59 (0.11) | 0.58 (0.11) | 0.58 (0.12) |
| Retropalatal lateral walls | 0.72 (0.09) | 0.72 (0.08) | 0.71 (0.09) | 0.72 (0.09) | 0.72 (0.09) |
| Retroglossal lateral walls | 0.71 (0.10) | 0.70 (0.09) | 0.70 (0.09) | 0.71 (0.09) | 0.71 (0.09) |
| Overall | 0.70 (0.10) | 0.70 (0.10) | 0.71 (0.10) | 0.70 (0.10) | 0.70 (0.10) |
Figure 3.
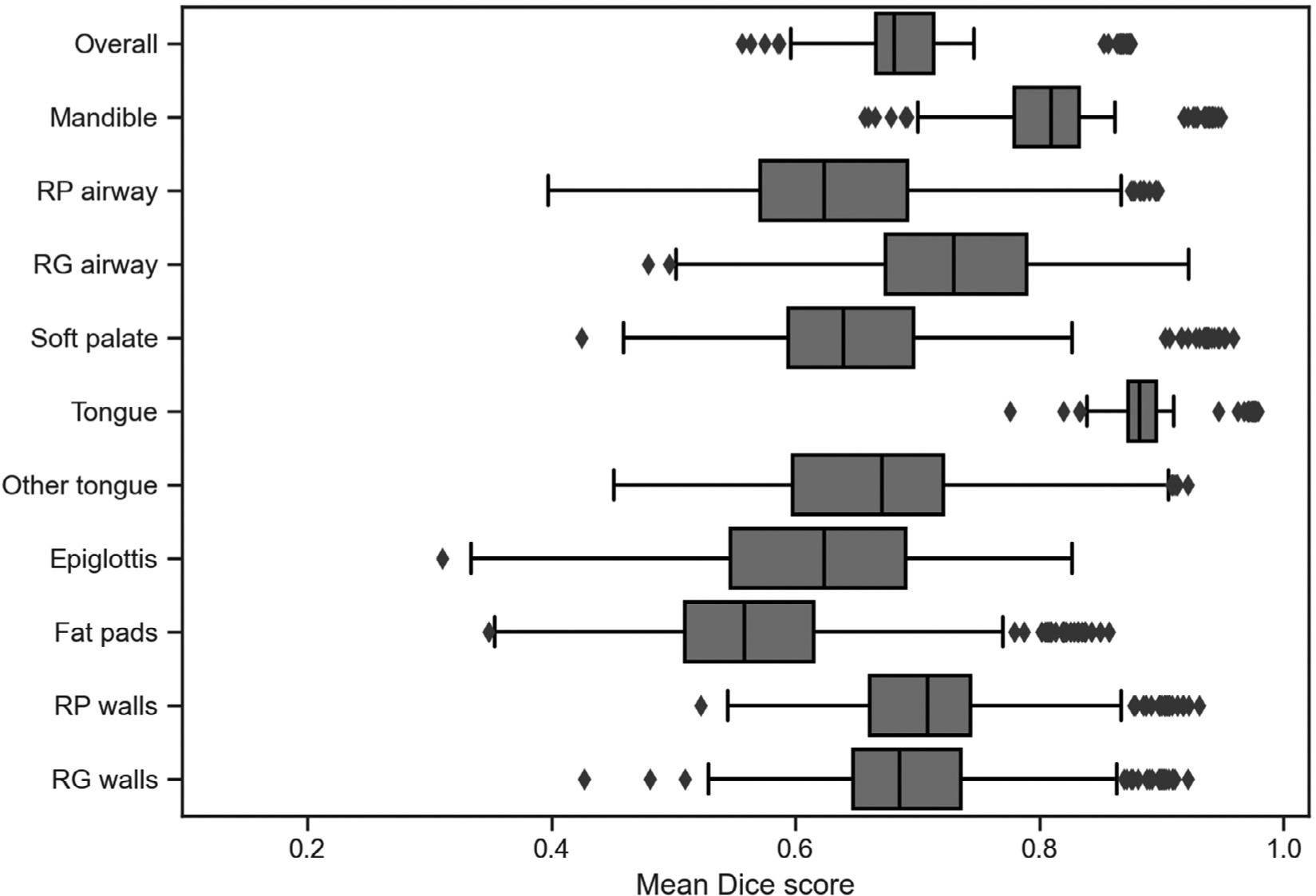
Fold-aggregated mean Dice Scores for each region of interest. Box and whisker plot for mean Dice Scores of each region of interest as well as of overall results. The diamonds represent outliers. The mean Dice scores are from the aggregated segmentation results of the cross-validations (n = 206). RG = retroglossal; RP = retropalatal.
In addition to cross-validated results, in the leave-study-out experiments, we found that mean segmentation accuracy was comparable to results obtained using all studies together — for each individual region of interest as well as for overall results (Table 3). In particular, the mean Dice score was highest for the model where Study two was excluded (0.71 ± 0.07) whereas the other three models had the same mean Dice score (Study one: 0.70 ± 0.07 and Studies three and four: 0.70 ± 0.10). Examples of segmentations from each leave-study-out model are shown in Figure 4.
TABLE 3.
Mean Dice Scores From Leave-Study-Out Validations When Tested on Each Excluded Batch (Along With One Standard Deviation): Study 1 (n = 31), Study 2 (n = 35), Study 3 (n = 64), and Study 4 (n = 76)
| Mean (Standard Deviation) Dice Scores from Leave-Study-Out Validations | ||||
|---|---|---|---|---|
| Structure | Model Trained With Study 1 Excluded | Model Trained With Study 2 Excluded | Model Trained With Study 3 Excluded | Model Trained With Study 4 Excluded |
| Mandible | 0.79 (0.06) | 0.81 (0.04) | 0.82 (0.07) | 0.82 (0.07) |
| Retropalatal airway | 0.63 (0.10) | 0.61 (0.10) | 0.63 (0.14) | 0.66 (0.12) |
| Retroglossal airway | 0.74 (0.08) | 0.74 (0.07) | 0.73 (0.10) | 0.74 (0.09) |
| Soft palate | 0.66 (0.09) | 0.65 (0.09) | 0.69 (0.13) | 0.67 (0.15) |
| Tongue | 0.89 (0.02) | 0.88 (0.03) | 0.89 (0.04) | 0.89 (0.04) |
| Other tongue | 0.69 (0.07) | 0.72 (0.06) | 0.69 (0.11) | 0.64 (0.13) |
| Epiglottis | 0.60 (0.12) | 0.66 (0.09) | 0.61 (0.12) | 0.56 (0.12) |
| Fat pads | 0.58 (0.07) | 0.59 (0.08) | 0.56 (0.14) | 0.58 (0.12) |
| Retropalatal lateral walls | 0.70 (0.07) | 0.71 (0.07) | 0.72 (0.09) | 0.72 (0.10) |
| Retroglossal lateral walls | 0.71 (0.07) | 0.70 (0.06) | 0.70 (0.10) | 0.70 (0.10) |
| Overall | 0.70 (0.07) | 0.71 (0.07) | 0.70 (0.10) | 0.70 (0.10) |
Figure 4.
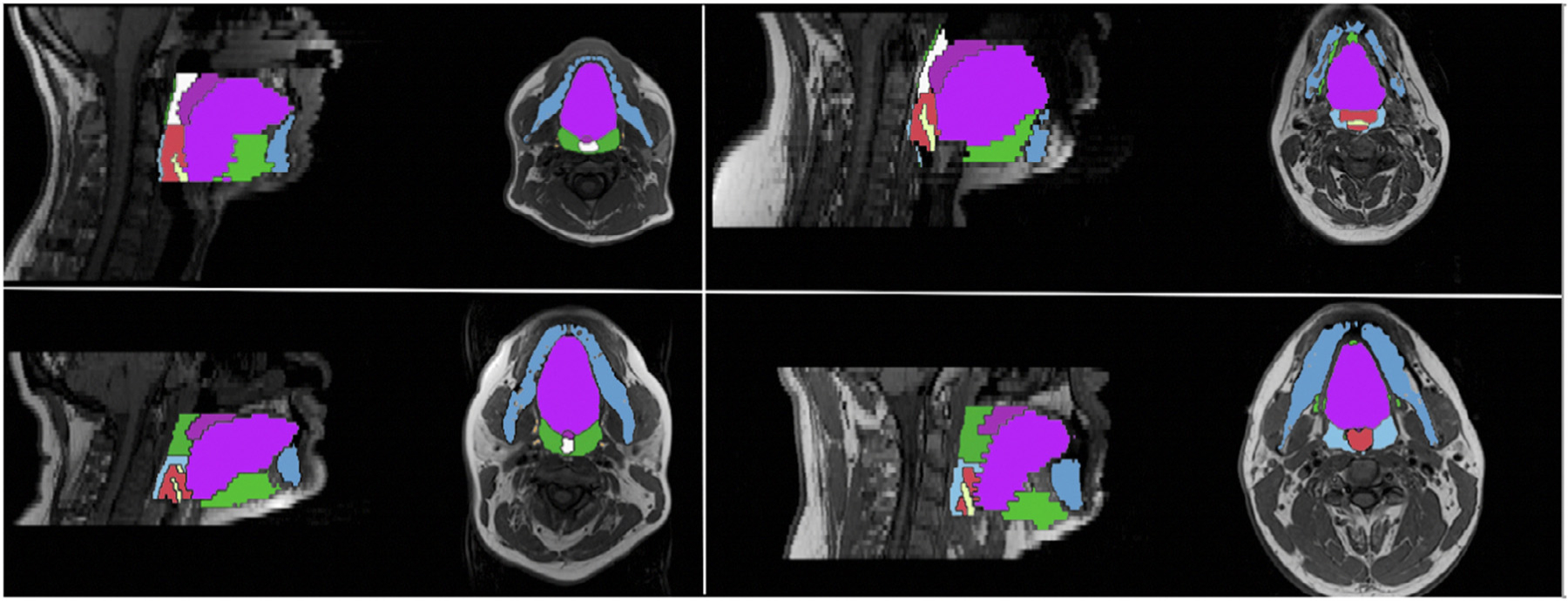
Automatically generated segmentations from each leave-study-out validation model. Four automatically generated segmentations from the leave-study-out validations, one for each excluded study model. Top left: Study 1, Top right: Study 2, Bottom left: Study 3, Bottom right: Study 4.
Agreement in Calculated Values Between Deep Learning and Manual Approaches
To understand the agreement and potential application of these methods, we compared the values of upper airway anatomy structure volumes and tongue fat volume ratio from manual (gold standard) and DL-based methods (see Table 4, Fig. 5–6). Regional volumes for automated segmentations from the cross-validation experiment showed a strong correlation with manually derived values (Fig 5 and Table 4) and generally strong agreement between the two methods based on Bland-Altman analysis (Fig 6). On average, we observed similar values using the two approaches, with very small standardized mean differences between the two techniques (| standardized mean differences | < 0.1) for all measures except the epiglottis; thus, statistically significant differences observed in Bland-Altman analyses were not clinically meaningful. There is some evidence of negative correlations between differences and averages on the two techniques (Table 4), suggesting more negative bias estimates for larger average values. However, we found that the limits of agreement lay within our a priori defined range of maximum acceptable differences for nearly all ROIs, except the epiglottis and the retropalatal airway. As indicated by the steeper slope of the linear fit compared to the slope of the identity line and the large standardized mean differences, there was a clear bias in the automatically segmented epiglottis volume. Automatically calculated tongue fat volume ratios V1 and V2 had a 0.85 and 0.96 correlation, respectively, with the values calculated from manual segmentation. The V2 ratio derived from the DL segmentations was not significantly different (p < 0.05) from the V2 ratio derived from the manual segmentations.
TABLE 4.
Comparison of the Volume of Anatomical Traits (in mm3) as Well as Comparison of Two Tongue Fat Volume Ratios Thresholded at Different Intensities From the Aggregated Cross-Validation Data (n = 206)
| Comparison of Anatomical Traits Derived from Manual and Deep Learning Methods | ||||||
|---|---|---|---|---|---|---|
| Structure | Manual | Deep Learning | Difference† | SMD‡ | Correlations§ | |
| Manual vs. DL | Mean vs. Difference | |||||
| Mandible | 55812.06 (12597.11) | 54624.29 (12521.55) | −1187.77 (1394.78)¶ | −0.094 | 0.99¶ | −0.05 |
| Retropalatal airway | 3474.71 (1840.78) | 3512.64 (1795.49) | 37.93 (237.48)¶ | 0.021 | 0.99¶ | −0.19¶ |
| Retroglossal airway | 6683.81 (3250.03) | 6682.33 (3141.60) | −1.48 (340.98) | 0.001 | >0.99¶ | −0.28¶ |
| Soft palate | 8568.51 (2763.32) | 8470.32 (2695.77) | −98.19 (358.92)¶ | −0.034 | 0.99¶ | −0.21¶ |
| Tongue | 84108.46 (18357.18) | 83966.11 (17746.98) | −142.34 (1615.92) | −0.006 | >0.99¶ | −0.38¶ |
| Other tongue | 29507.51 (10716.46) | 29100.40 (10583.58) | −407.12 (960.80)¶ | −0.040 | >0.99¶ | −0.13 |
| Epiglottis | 1517.73 (829.25) | 2023.98 (1024.29) | 506.26 (271.34)¶ | 0.612 | 0.98¶ | 0.73¶ |
| Fat pads | 4446.24 (1775.67) | 4388.04 (1740.38) | −58.21 (127.93)¶ | −0.031 | >0.99¶ | −0.31¶ |
| Retropalatal lateral walls | 11146.57 (4196.97) | 11024.67 (4115.13) | −121.91 (459.52)¶ | −0.028 | 0.99¶ | −0.17¶ |
| Retroglossal lateral walls | 9727.31 (3832.54) | 9625.61 (3735.65) | −101.70 (245.50)¶ | −0.026 | >0.99¶ | −0.38 |
| Tongue Fat Ratio (V1) | 0.225 (0.06) | 0.228 (0.06) | 0.003 (0.032) | 0.057 | 0.85¶ | −0.17¶ |
| Tongue Fat Ratio (V2) | 0.253 (0.07) | 0.258 (0.06) | 0.005 (0.019)¶ | 0.076 | 0.96¶ | −0.18¶ |
Subject-specific difference calculated as deep learning (DL) minus manual.
Standardized mean difference (SMD) calculated as the mean difference divided by standard deviation from manual measurement – values of 0.2, 0.5, and 0.8 represent small, medium and large differences, respectively (21).
Pearson correlation coefficients between manual and DL based values or between average and difference on the two techniques.
p < 0.05 from paired t-test (for difference) or correlation analysis.
Figure 5.
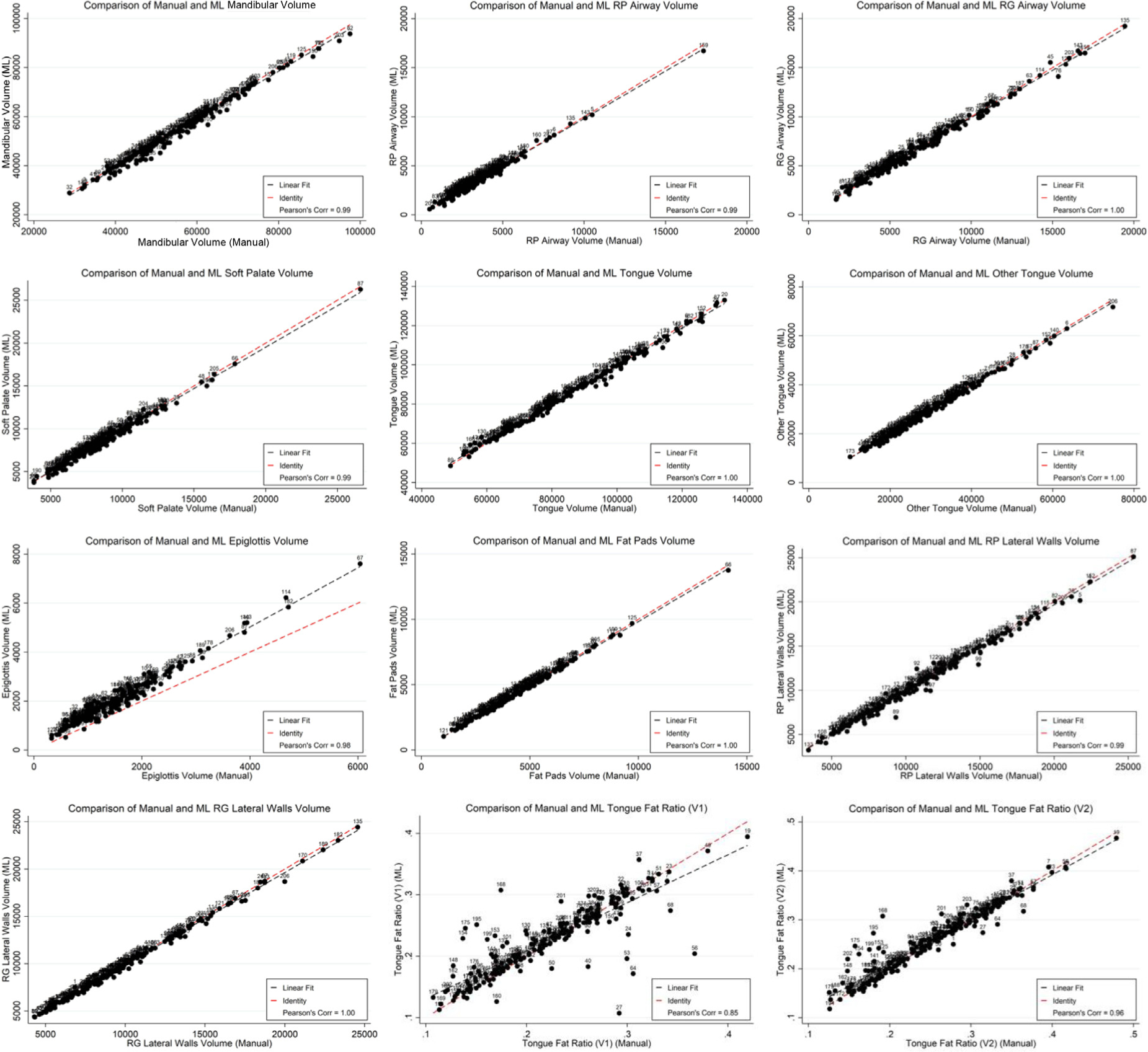
Spearman correlation plots between anatomical traits derived from manual and deep learning methods. Spearman correlation plots between each of 10 manually segmented and deep learning-segmented ROI volumes (in mm3) as well as 2 correlation plots of the relation between each manually derived adjusted tongue fat volume ratio and deep learning-derived adjusted tongue fat volume ratio. The black dashed line represents the calculated correlation between the automated and manual segmentation volumes whereas the red dashed line represents the identity line. The deep learning-derived ratios and ROI volumes are calculated from the aggregated segmentation results of the cross-validation experiment (n = 206). ML = machine learning; RG = retroglossal; RP = retropalatal; V1 = voxel-weighted at max fat pad intensity; V2 = voxel-weighted at 99th percentile fat pad intensity.
Figure 6.
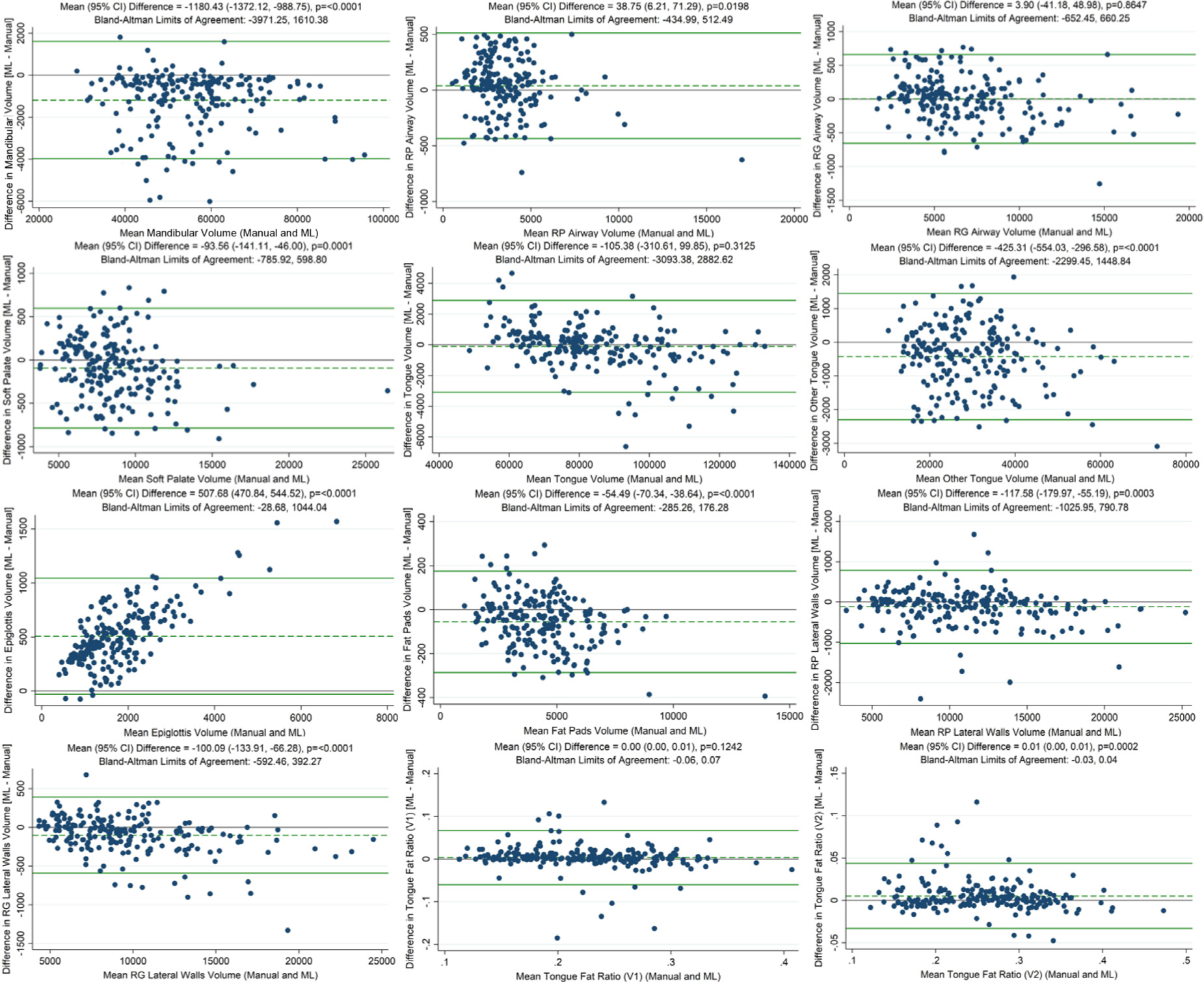
Bland-Altman plots of the relation between anatomical traits derived from manual and deep learning methods. Bland-Altman plots of the relation between each of 10 manually segmented and deep learning-segmented ROI volumes (in mm3) as well as 2 Bland-Altman plots of the relation between both manually derived adjusted tongue fat volume ratios and both deep learning-derived adjusted tongue fat volume ratios. The dark green lines indicated the upper and lower limits of agreement whereas the dashed green line represents the mean difference. The deep learning-derived ratios and ROI volumes are from the aggregated results of the cross-validation experiment (n = 206). ML = machine learning; RG = retroglossal; RP = retropalatal; V1 = voxel-weighted at max fat pad intensity; V2 = voxel-weighted at 99th percentile fat pad intensity.
DISCUSSION
We presented a fully automated method to segment 10 upper airway structures that are anatomic risk factors for OSA and to derive the tongue fat biomarker from T1-weighted MRI scans using a dataset of 206 unique subjects from four different clinical studies. To the best of our knowledge, this is the first study that uses DL techniques to address segmentation of the important pharyngeal soft tissue anatomic risk factors for OSA. The proposed algorithm was built on a generic CNN architecture that integrates components designed based on recent advances in neuroimage analysis using DL. (4,5) Overall, in our evaluation experiments using four-fold cross-validation, our model obtained high accuracy and agreement, indicating its potential for application in the study of large imaging datasets to understand the pathogenesis of OSA.
Inter-rater agreement has been established for the MR upper airway measures between human scorers (22); using these established criteria, nearly all the Dice scores reported in this paper fall within the “almost perfect” (0.80–0.99) and “substantial” (0.61–0.80) ranges. The tongue region was the region with highest mean segmentation accuracy, with an average (standard deviation) Dice score of 0.89 (0.04), whereas the fat pads region had the lowest mean accuracy, with an average (standard deviation) Dice score of 0.58 (0.12). In our four-fold cross-validation experiment, we had identical 95% CIs for overall Dice scores across three folds and a lack of significant volume differences (p < 0.05) in multiple structures that are more difficult to segment due to small size or lack of image contrast in region boundaries, such as the mandible, retroglossal airway, and soft palate. Just as importantly, most Dice scores were within the same inter-rater agreement ranges between subjects and datasets. Taken together, this suggests that our algorithm is not affected by image and cohort variations.
A primary goal of quantitative image analysis is to derive imaging biomarkers that can be clinically used to determine anatomic risk factors for OSA, including an enlarged volume of the tongue, lateral walls, soft palate, increased tongue fat volume ratio and a narrow upper airway. (23) Previous analyses were limited to relatively small samples, mainly due to the difficulties and effort required for manual segmentation of upper airway structures. In our comparisons against manual calculations used as the ground-truth, DL derived volumes for nearly all pharyngeal structures were highly correlated (p ≥ 0.98), not significantly different from manual segmentation (p < 0.05), and within the limits of maximum acceptable differences. In addition, a completely automated tongue fat volume ratio obtained a very high correlation (p = 0.96) and was not significantly different (p < 0.05) relative to manual segmentation. Surprisingly, the agreement between automated and manual ratios was high across all cases — even for cases that obtained relatively low Dice scores in tongue segmentation. This result indicates that even when the automated and the ground truth segmentations do not completely agree, the final biomarker that is derived using each approach is similar. This may indicate the robustness of the final biomarker to variations due to scanners, subjects, or methodology used for segmentation. Our results indicate that the proposed approach could have important practical utility for automatically deriving imaging biomarkers in large scale research or clinical studies. Many patients undergo MRI of the neck and head for clinical reasons unrelated to sleep apnea. We could potentially apply our DL algorithms in the background to automatically segment (calculate volumes) of the soft tissues surrounding the upper airway to identify patients with anatomic risk factors for sleep apnea. Such patients could then be screened for sleep apnea.
A concern for DL methods is generalizability, e.g., its applicability to imaging datasets never seen during training. Using a heterogeneous training set that captures the expected variations in input images is a commonly used strategy for obtaining more robust results. In this study, we combined four different datasets and performed experiments using careful cross-validation to test the generalizability of the final trained model. To further test our method’s robustness, we performed leave-study-out cross-validation. As such, we were able to test the potential of our model to work on datasets with different acquisition methods (leave-study-out validations). The accuracy in leave-study-out experiments was comparable to the accuracy of models with four-fold cross-validations. The current results are promising, especially for application to larger-scale studies with similar imaging approaches; nevertheless, further training on patients with more variable morphology in their upper airway would be valuable to improve generalizability and ultimately facilitate incorporating these algorithms more routinely in clinical practice. In particular, a range of scans from a diverse patient population (i.e., in age and in upper airway morphology) will allow the model to generalize with greater accuracy.
Although our model obtains clinically acceptable agreement for nearly all pharyngeal structures, this study still has some limitations. The epiglottis segmentation had an obvious bias, with the volumes being overestimated. Since our model minimized the error for the joint set of all structures, relatively smaller structures, such as the epiglottis, retropalatal airway, and fat pads, would be expected to yield higher error rates. Moreover, while we used a unique multi-study dataset with manual ground-truth segmentations and with a moderately large sample size, application of the proposed method on a larger clinical scale would require further training using a larger, more heterogeneous sample set. We plan to address these challenges in future work using semi-supervised learning strategies. Another limitation is that our method is applied on T1-weighted images, and we need to perform more validation studies on samples acquired from a greater variety of scanners. Using multi-modal imaging may improve segmentation accuracy and increase potential for clinical usage in a real-world setting. Our network architecture is readily adaptable to a multi-modal setting. We are working on collecting multi-modal data for extending our method to multi-modal scans in future work.
In conclusion, the results of our quantitative evaluations indicate high spatial accuracy and volumetric agreement with manual standards and demonstrate the potential of DL-based segmentation for automating the segmentation and quantification of upper airway anatomic risk factors for OSA in large-scale studies. This, in turn, could facilitate several research and clinical applications, including genetic association studies in electronic health records with linked genetic data or potential for screening for obstructive sleep apnea by applying this DL algorithm to clinically obtained images and quantifying relevant risk factors, including the volume of the tongue and tongue fat.
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abbreviations:
- OSA
obstructive sleep apnea
- MRI
magnetic resonance imaging
- MR
magnetic resonance
- DL
deep learning
- CNN
convolutional neural network
- ROI
region of interest
- V1
voxel-weighted at max fat pad intensity
- V2
voxel-weighted at 99th percentile fat pad intensity
Footnotes
SUPPLEMENTARY MATERIALS
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.acra.2022.04.023.
REFERENCES
- 1.Benjafield AV, Ayas NT, Eastwood PR, et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med 2019; 7(8):687–698. doi: 10.1016/s2213-2600(19)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patil SP, Schneider H, Schwartz AR, et al. Adult obstructive sleep apnea. Chest 2007; 132(1):325–337. doi: 10.1378/chest.07-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwab RJ, Pasirstein M, Pierson R, et al. Identification of upper airway anatomic risk factors for obstructive sleep apnea with volumetric magnetic resonance imaging. Am J Respir Crit Care Med 2003; 168(5):522–530. doi: 10.1164/rccm.200208-866oc. [DOI] [PubMed] [Google Scholar]
- 4.Wang SH, Keenan BT, Wiemken A, et al. Effect of weight loss on upper airway anatomy and the apnea–Hypopnea index. The importance of tongue fat. Am J Respir Crit Care Med 2020; 201(6):718–727. doi: 10.1164/rccm.201903-0692oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim AM, Keenan BT, Jackson N, et al. Tongue fat and its relationship to obstructive sleep apnea. Sleep 2014; 37(10):1639–1648. doi: 10.5665/sleep.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521 (7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Kleesiek J, Urban G, Hubert A, et al. Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. NeuroImage 2016; 129:460–469. doi: 10.1016/j.neuroimage.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal 2017; 35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Yu Y, Wang Q, et al. Upper airway segmentation based on the attention mechanism of weak feature regions. IEEE Access 2021; 9:95372–95381. doi: 10.1109/access.2021.3094032. [DOI] [Google Scholar]
- 10.Park J, Hwang JJ, Ryu J, et al. Deep learning based airway segmentation using key point prediction. Appl Sci 2021; 11(8):3501. doi: 10.3390/app11083501. [DOI] [Google Scholar]
- 11.Alsufyani NA, Flores-Mir C, Major PW. Three-dimensional segmentation of the upper airway using cone beam CT: a systematic review. Dentomaxillofac Radiol 2012; 41(4):276–284. doi: 10.1259/dmfr/79433138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shahid ML, Chitiboi T, Ivanovska T, et al. Automatic MRI segmentation of para-pharyngeal fat pads using interactive visual feature space analysis for classification. BMC Med Imaging 2017; 17(1). doi: 10.1186/s12880-017-0179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanovska T, Dober J, Laqua R, et al. Pharynx segmentation from MRI data for analysis of sleep related disorders. Adv Vis Comput 2013: 20–29. doi: 10.1007/978-3-642-41914-0_3. [DOI] [Google Scholar]
- 14.Xie L, Udupa JK, Tong Y, et al. Automatic upper airway segmentation in static and dynamic MRI via anatomy-guided Convolutional Neural Networks. Med Phys 2021; 49(1):324–342. doi: 10.1002/mp.15345. [DOI] [PubMed] [Google Scholar]
- 15.Doshi J, Erus G, Habes M, et al. DeepMRSeg: a convolutional deep neural network for anatomy and abnormality segmentation on MR images. arXiv. Preprint posted online July 3, 2019. Available at: https://arxiv.org/abs/1907.02110. Accessed 01 Mar 2022. [Google Scholar]
- 16.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sci 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 17.He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. doi: 10.1109/cvpr.2016.90. [DOI] [Google Scholar]
- 18.Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-ResNet and the impact of residual connections on learning. arXiv. Preprint posted online February 23, 2016. Available at: https://arxiv.org/abs/1602.07261. Accessed 01 Mar 2022. [Google Scholar]
- 19.Humbert IA, Reeder SB, Porcaro EJ, et al. Simultaneous estimation of tongue volume and fat fraction using ideal-FSE. J Magn Reson Imaging 2008; 28(2):504–508. doi: 10.1002/jmri.21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin Bland J, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986; 327 (8476):307–310. doi: 10.1016/s0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 21.Cohen J Statistical power analysis for the behavioral sciences. New York, NJ: Psychology Press, 2009. [Google Scholar]
- 22.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012; 22(3):276–282. [PMC free article] [PubMed] [Google Scholar]
- 23.Pro Schwab RJ. Sleep apnea is an anatomic disorder. Am J Respir Crit Care Med 2003; 168(3):270–271. doi: 10.1164/rccm.2305014. [DOI] [PubMed] [Google Scholar]


