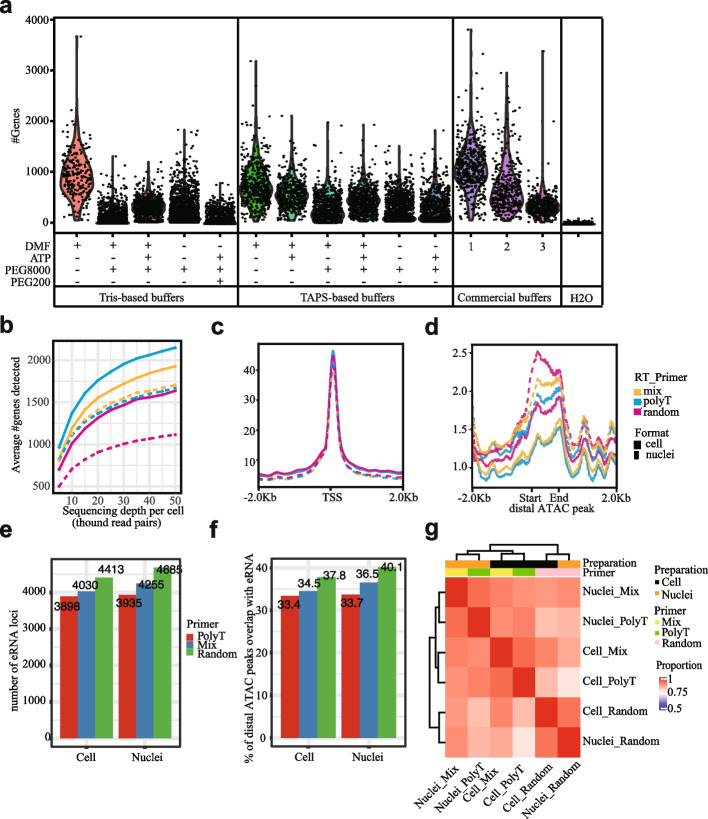
Fig. 2.
Optimization and expanded application of FIPRESCI. a Violin plot showing sensitivity in FIPRESCI generated with a set of TN5 tagmentation buffers. Each dot represents a single cell. Y-axis is the number of genes detected. b Sensitivity of gene detection in three reverse transcription primer conditions in FIPRESCI single-cell RNA-seq (solid line) and single-nucleus RNA-seq (dotted line) across sequencing depth. c Distribution of reads from three reverse transcription primer conditions in FIPRESCI scRNA-seq and snRNA-seq around annotated TSS. d Distribution of reads from three reverse transcription primer conditions in FIPRESCI scRNA-seq and snRNA-seq around enhancer center. e Histogram showing the number of eRNA loci identified in three reverse transcription primer conditions in FIPRESCI scRNA-seq and snRNA-seq. f Histogram showing the percentage of distal ATAC peaks which are overlap with eRNA loci in three reverse transcription primer conditions in FIPRESCI scRNA-seq and snRNA-seq. g Heatmap showing pairwise correlations, hierarchical clustering for the eRNA profile across different reverse transcription primers, and preparation methods using FIPRESCI